Abstract
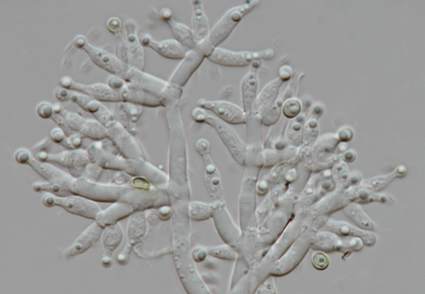
Trichoderma species are distributed worldwide and they play an important role in plant protection, growth promotion and also as biocontrol agents of plant diseases. In this study, a new species of Trichoderma was isolated from Azadirachta indica rhizosphere soils in Zhaoqing City, Guangdong Province, China. Based on the morphology and combined multigene phylogeny of nuclear internal transcribed spacer of rDNA region (ITS), translation elongation factor 1 alpha (tef1-α) and RNA polymerase second largest subunit (rpb2), a new Trichoderma species belongs to the Harzianum-clade was identified, namely Trichoderma azadirachtae sp. nov. Distinctions between the new species and its close relatives are discussed in detail.
References
- Bruen, T.C., Philippe, H. & Bryant, D. (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172 (4): 2665–2681. https://doi.org/10.1534/genetics.105.048975
- Bustamante, D.E., Calderon, M.S., Leiva, S., Mendoza, J.E., Arce, M. & Oliva, M. (2021) Three new species of Trichoderma in the Harzianum and Longibrachiatum lineages from Peruvian cacao crop soils based on an integrative approach. Mycologia 113 (5): 1056–1072. https://doi.org/10.1080/00275514.2021.1917243
- Cai, F. & Druzhinina, I.S. (2021) In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Diversity 107: 1–69. https://doi.org/10.1007/s13225-020-00464-4
- Cao, Z.J., Zhao, J., Liu, Y., Wang, S.X., Zheng, S.Y. & Qin, W.T. (2024) Diversity of Trichoderma species contaminating substrates of Lentinula edodes in China and their interaction evaluation. Frontiers in Microbiology 14: 1288585. https://doi.org/10.3389/fmicb.2023.1288585
- Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. https://doi.org/10.1080/00275514.1999.12061051
- Chaverri, P., Branco-Rocha, F., Jaklitsch, W., Gazis, R., Degenkolb, T. & Samuels, G.J. (2015) Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107: 558–590. https://doi.org/10.3852/14-147.
- De Silva, N.I., Brooks, S., Lumyong, S. & Hyde, K.D. (2019) Use of endophytes as biocontrol agents. Fungal Biology Reviews 33 (2): 133–148. https://doi.org/10.1016/j.fbr.2018.10.001
- Druzhinina, I.S., Kopchinskiy, A.G. & Kubicek, C.P. (2006) The first 100 Trichoderma species characterized by molecular data. Mycoscience 47 (2): 55–64. https://doi.org/10.1007/s10267-006-0279-7
- Gu, X., Wang, R., Sun, Q., Wu, B. & Sun, J.Z. (2020) Four new species of Trichoderma in the Harzianum clade from northern China. MycoKeys 73: 109–132. https://doi.org/10.3897/mycokeys.73.51424
- Hall, T. (2006) Bioedit. Raleigh, NC: Department of Microbiology, North Carolina State University. Available from: http://www.mbio.ncsu.edu/BioEdit/page2.html (accessed 31 October 2024)
- Hyde, K.D., McKenzie, EHC. & Koko, TW. (2011) Towards incorporating anamorphic fungi in a natural classification - checklist and notes for 2010. Mycosphere 2 (1): 1–88. [https://mycosphere.org/pdf/MC2_1_No1.pdf]
- Jaklitsch, W.M., Komon, M., Kubicek, C.P. & Druzhinina, I.S. (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97: 1365–1378. https://doi.org/10.1080/15572536.2006.11832743
- Kaewchai, S., Soytong, K. & Hyde, K.D. (2009) Mycofungicides and fungal biofertilizers. Fungal Diversity 38: 25–50. [https://api.semanticscholar.org/CorpusID:53870012]
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: Multiple sequence align ment, interactive sequence choice and visualization. Briefings in Bioinformatics 20 (4): 1160–1166. https://doi.org/10.1093/bib/bbx108
- Liu, X.F., Tibpromma, S., Hughes, A.C., Chethana, K., Wijayawardene, N.N., Dai, D.Q., Du, T.Y., Elgorban, A.M., Stephenson, S.L., Suwannarach, N., Xu, J.C., Lu, L., Xu, R.F., Maharachchikumbura, S., Zhao, C.L., Bhat, D.J., Sun, Y.M., Karunarathna, S.C. & Mortimer, P.E. (2023) Culturable mycota on bats in central and southern Yunnan Province, China. Mycosphere 14 (1): 497–662. https://doi.org/10.5943/mycosphere/14/1/7
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Luo, M., Chen, Y., Huang, Q.R., Huang, Z.X., Song, H.D. & Dong, Z.Y. (2023) Trichoderma koningiopsis Tk905: an efficient biocontrol, induced resistance agent against banana Fusarium wilt disease and a potential plant-growth-promoting fungus. Frontiers in Microbiology 14: 1301062. https://doi.org/10.3389/fmicb.2023.1301062
- Maharachchikumbura, S.S.N., Chen, Y., Ariyawansa, H.A., Hyde, K.D., Haelewaters, D., Perera, R.H., Samarakoon, M.C., Wanasinghe, D.N., Bustamante, D.E., Liu, J., Lawrence, D.P., Cheewangkoon, R. & Stadler, M. (2021) Integrative approaches for species delimitation in Ascomycota. Fungal Diversity 109: 155–179. https://doi.org/10.1007/s13225-021-00486-6
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). Institute of Electrical and Electronics Engineers, pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Nuangmek, W., Aiduang, W., Kumla, J., Lumyong, S. & Suwannarach, N. (2021) Evaluation of a Newly Identified Endophytic Fungus, Trichoderma phayaoense for Plant Growth Promotion and Biological Control of Gummy Stem Blight and Wilt of Muskmelon. Frontiers in Microbiology 12: 634772. https://doi.org/10.3389/fmicb.2021.634772
- Nylander, J.A.A. (2004) MrModeltest 2.0. Program distributed by the author. Uppsala, Sweden: Evolutionary Biology Centre Uppsala University. [http://www.abc.se/~nylander/mrmodeltest2/mrmodeltest2.html]
- Overton, B.E., Stewart, E.L. & Geiser, D.M. (2006) Taxonomy and phylogenetic relationships of nine species of Hypocrea with anamorphs assignable to Trichoderma section Hypocreanum. Studies in Mycology 56 (1): 39–65. https://doi.org/10.3114/sim.2006.56.02
- Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 (12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
- Rosado, J.P., Palacios, C.S., Cumbicus, J.J., Puga, E.P., Sabando, G. & Olalla, L.A. (2024) In vitro evaluation of the inhibitory capacity of three Trichoderma isolates on Ralstonia solanacearum. Bionatura 9 (1): 6. http://dx.doi.org/10.21931/rb/2024.09.01.6
- Samuels, G.J., Dodd, S.L., Lu, B.S., Petrini, O, Schroers, H.J. & Druzhinina, I.S. (2006) The Trichoderma koningii aggregate species. Studies in mycology 56 (1): 67–133. https://doi.org/10.3114/sim.2006.56.03
- Silvestro, D. & Michalak, I. (2012) RaxmlGUI: a Graphical Front-End for RAxML. Organisms Diversity & Evolution 12: 335–337. https://doi.org/10.1007/s13127-011-0056-0
- Species Fungorum. (2024) Available from: http://www.speciesfungorum.org/Names/Names.asp (accessed 1 February 2024)
- Stamatakis, A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9): 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Stielow, J.B., Lévesque, C.A., Seifert, K.A., Meyer, W., Iriny, L., Smits, D., Renfurm, R., Verkley, G.J., Groenewald, M., Chaduli, D., Lomascolo, A., Welti, S., Lesage-Meessen, L., Favel, A., Al-Hatmi, A.M., Damm, U., Yilmaz, N., Houbraken, J., Lombard, L., Quaedvlieg, W., Binder, M., Vaas, L.A., Vu, D., Yurkov, A., Begerow, D., Roehl, O., Guerreiro, M., Fonseca, A., Samerpitak, K., van, Diepeningen, A.D., Dolatabadi, S., Moreno, L.F., Casaregola, S., Mallet, S., Jacques, N., Roscini, L., Egidi, E., Bizet, C., Garcia-Hermoso, D., Martín, M.P., Deng, S., Groenewald, J.Z., Boekhout, T., de Beer, Z.W., Barnes, I., Duong, T.A., Wingfield, M.J., de Hoog, G.S., Crous, P.W., Lewis, C.T., Hambleton, S., Moussa, T.A., Al-Zahrani, H.S., Almaghrabi, O.A., Louis-Seize, G., Assabgui, R., McCormick, W., Omer, G., Dukik, K., Cardinali, G., Eberhardt, U., de, Vries, M. & Robert, V. (2015) One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35: 242–263. https://doi.org/10.3767/003158515X689135
- Swofford, D.L. (2002) PAUP: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0b10. Sinauer Associates, Sunderland. https://doi.org/10.1111/j.0014-3820.2002.tb00191.x
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sánchez-García, M., Goto, B.T., Saxena, R.K., Erdoğdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., Błaszkowski, J., Boonyuen, N., da Silva, G.A., de Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Mešić, A., Monteiro, J.S., Oehl, F., Pawłowska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro, I., Tang, L.Z., Stadler, M., Yurkov, A. & Thines, M. (2022) Outline of fungi and fungus-like taxa – 2021. Mycosphere 13 (1): 53–453. https://doi.org/10.5943/mycosphere/13/1/2
- Ye, C.W., Jing, T.T., Sha, Y.R., Mo, M.H. & Yu, Z.F. (2023) Two new Trichoderma species (Hypocreales, Hypocreaceae) isolated from decaying tubers of Gastrodia elata. MycoKeys 99: 187–207. https://doi.org/10.3897/mycokeys.99.109404
- Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20 (1): 348–355. https://doi.org/10.1111/1755-0998.13096
- Zhao, R., Mao, L.J. & Zhang, C.L. (2023) Three new species of Trichoderma (Hypocreales, Hypocreaceae) from soils in China. MycoKeys 97: 21–40. https://doi.org/10.3897/mycokeys.97.101635
- Zheng, H., Qiao, M., Lv, Y.F., Du, X., Zhang, K.Q. & Yu, Z.F. (2021) New species of Trichoderma isolated as endophytes and saprobes from Southwest China. Journal of Fungi 7: 467. https://doi.org/10.3390/jof7060467