Abstract
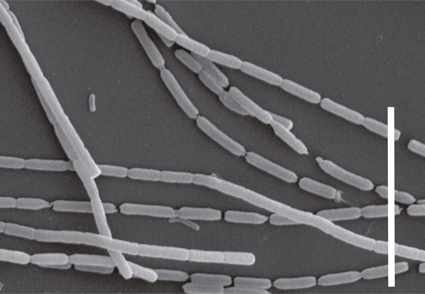
Two Leptolyngbya-like cyanobacterial strains were isolated from two marine benthic environments on the Brazilian coast. These strains were cultured and characterized based on their morphology, molecular, and ultrastructural data. The two taxa were identified mainly by 16S rRNA gene phylogeny and 16S-23S ITS secondary structures since their morphology is similar to members of Leptolyngbya senso lato. The phylogenetic analysis indicated that our strains belong to Euryhalinema genus (Leptolyngbyaceae), with one strain close to type species E. mangrovii AP9F (MK402979), and the other described as a new species, E. epiphyticum sp. nov. Morphologically, both strains form green mats, had trichomes without sheath, and their thylakoid disposition is the same as described for Leptolyngbyaceae. The secondary structures Box B and D1-D1’ of the internal transcribed spacer (16S–23S ITS) also corroborated our proposal of the new species E. epiphyticum. These findings constitute the first description of a new-to-science species for this genus outside Indian marine environments. Also, it expands the knowledge on Euryhalinema systematic.
References
<p>Becerra-Absalón, I., Johansen, J.R., Muñoz-Martín, M.A. & Montejano, G. (2018) <em>Chroakolemma gen. nov</em>. (Leptolyngbyaceae, Cyanobacteria) from soil biocrusts in the semi-desert Central Region of Mexico. <em>Phytotaxa </em>367: 201–218. https://doi.org/10.11646/phytotaxa.367.3.1</p>
<p>Birnboim, H.C. & Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. <em>Nucleic Acids Research</em> 7: 1513–1518. https://doi.org/10.1093/nar/7.6.1513</p>
<p>Caires, T.A., Sant’Anna, C.L., Nunes, J.M.C. (2019) Biodiversity of benthic filamentous cyanobacteria in tropical marine environments of Bahia State, Northeastern Brazil. <em>Brazilian Journal of Botany</em> 42: 149–170. https://doi.org/10.1007/s40415-019-00517-2</p>
<p>Castenholz, R.W. (1988) Culturing methods for cyanobacteria. <em>Methods Enzymol</em> 167: 68–93. https://doi.org/10.1016/0076-6879(88)67006-6</p>
<p>Chakraborty, S., Maruthanayagam, V., Achari, A., Pramanik, A., Jaisankar, P. & Mukherjee, J. (2019) <em>Euryhalinema mangrovii</em> <em>gen. nov.</em>, <em>sp. nov.</em> and <em>Leptoelongatus litoralis</em> <em>gen. nov.,</em> <em>sp. nov.</em> (Leptolyngbyaceae) isolated from an Indian mangrove forest. <em>Phytotaxa</em> 422 (1): 58–74. https://doi.org/10.11646/phytotaxa.422.1.4</p>
<p>Dvořák, P., Jahodářová, E., Hašler, P., Gusev, E., Poulíčková, A. (2015) A new tropical cyanobacterium <em>Pinocchia polymorpha gen. et sp. nov.</em> derived from the genus <em>Pseudanabaena. Fottea</em> 15 (1): 113–120. https://doi.org/10.5507/fot.2015.010</p>
<p>Iteman, I., Rippka, R., de Marsac, N.T., Hergman, M. (2000) Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. <em>Microbiology</em> 146: 1275–1286. https://doi.org/10.1099/00221287-146-6-1275</p>
<p>Jacinavicius, F.R., Gama, W.A.Jr., Azevedo, M.T.P. & Sant’Anna, C.L. (2012) Manual para cultivo de cianobactérias. Publicações Online do Instituto de Botânica de São Paulo, São Paulo, 32 pp.</p>
<p>Karnovsky, M.J. (1965) A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. <em>Journal of Cell Biology </em>27: 1A–149A.</p>
<p>Komárek, J. (2007) Phenotype diversity of the cyanobacterial genus <em>Leptolyngbya</em> in the maritime Antartic. <em>Polish Polar Research</em>, 20 (3): 211–231.</p>
<p>Komárek, J. & Anagnostidis, K. (2005) Cyanoprokaryota-2. Teil/2nd part: Oscillatoriales.<em> In: </em>Büdel, B., Gärtner, G., Krienitz, L. & Schagerl, M. (Eds.) Süsswasserflora von Mitteleuropa 19 ⁄ 778 2. <em>Elsevier/Spektrum</em>, Heidelberg, pp. 1–759.</p>
<p>Komárek, J., Kaštovský, J., Mareš, J., Johansen, J.R. (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. <em>Preslia </em>86: 295–335.</p>
<p>Konstantinou, D., Voultsiadou, E., Panteris, E., Zervou, S-K., Hiskia, A., Gkelis, S. (2019) <em>Leptothoe</em>, a new genus of marine cyanobacteria (Synechococcales) and three new species associated with sponges from the Aegean Sea. <em>J</em>. <em>Phycol</em>. 55: 882–897. https://doi.org/10.1111/jpy.12866</p>
<p>Lane, D.J. (1991) 16S/23S rRNA sequencing. In <em>Nucleic Acid Techniques in Bacterial Systematics</em> (Stackebrandt, E., Goodfellow, M., editors). John Wiley and Sons, Chichester, United Kingdom, pp. 115–175.</p>
<p>Lukesová, A., Johansen, J.R., Martin, M.P., Casamatta, D.A. (2009) <em>Aulosira bohemensis</em> <em>sp. nov.</em>: further phylogenetic uncertainty at the base of Nostocales (Cyanobacteria). <em>Phycologia</em> 48: 118–129. https://doi.org/10.2216/08-56.1</p>
<p>Mai, T., Johansen, J., Pietrasiak, N., Bohunická, M., Martin, M.P. (2018) Polyphasic characterization of four species of <em>Pseudanabaena </em>(Oscillatoriales, Cyanobacteria) from China and insights into polyphyletic divergence within the <em>Pseudanabaena </em>genus. <em>Phytotaxa</em>, 365 (1): 001–059.</p>
<p>Mau, B., Newton, M.A., Larget, B., 1999. Bayesian phylogenetic inference via Markov chain Monte Carlo methods. <em>Biometrics</em> 55: 1–12. https://doi.org/10.1111/j.0006-341X.1999.00001.x</p>
<p>Neilan, B., A., Jacobs, D., DelDot, T., Blackall, L.L., Hawkins, P.R., Cox, P.T., Goodman, A.E. (1997) rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus <em>Microcystis</em>. <em>International journal of systematic bacteriology</em> 47: 693–697. https://doi.org/10.1099/00207713-47-3-693</p>
<p>Nylander, J.A.A. (2008) MrModeltest 2.3. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University.</p>
<p>Osorio–Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Kováčik, L., Martin, M.P. Johansen, J.R. (2014) Seven new species of <em>Oculatella</em> (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. <em>European journal of phycology</em> 49: 450–470. https://doi.org/10.1080/09670262.2014.976843</p>
<p>Posada, D. & Crandall, K. (1998) MODELTEST: Testing the Model of DNA Substitution. <em>Bioinformatics</em> 14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817</p>
<p>R Core Team. (2017) <em>R: A language and environment for statistical computing.</em> Vienna: R Foundation for Statistical Computing. [http://www.R-project.org/]</p>
<p>Radzi, R., Muangmai, N., Broady, P., Omar, W.M.W., Lavoue, S., Convey, P. & Merican, F. (2019) <em>Nodosilinea signiensis</em> <em>sp. nov.</em> (Leptolyngbyaceae, Synechococcales), a new terrestrial cyanobacterium isolated from mats collected on Signy Island, South Orkney Islands, Antarctica. <em>PLoS ONE</em> 14 (11): e0224395. https://doi.org/10.1371/journal.pone.0224395</p>
<p>Rippka, R., Deruelles, J., Waterbury, J.B., Herdman, M. & Stanier, R.Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. <em>Journal of General Microbiology</em> 111: 1–61. https://doi.org/10.1099/00221287-111-1-1</p>
<p>Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. <em>Syst</em>. <em>Biol</em>. 61: 539– 847. https://doi.org/10.1093/sysbio/sys029</p>
<p>Sambrook, J.F. & Russell, D.W. (2001) <em>Molecular cloning: a laboratory manual</em>. Cold Spring Harbor Laboratory Press, New York, 2100 pp.</p>
<p>Sciuto, K. & Moro, I. (2016) Detection of the new cosmopolitan genus <em>Thermoleptolyngbya </em>(Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S-23S ITS region. <em>Molecular Phylogenetics and Evolution</em> 105: 15–35. https://doi.org/10.1016/j.ympev.2016.08.010</p>
<p>Sciuto, K., Moschin, E. & Moro, I. (2017) Cryptic cyanobacterial diversity in the giant cave (Trieste, Italy): the new genus <em>Timaviella </em>(Leptolyngbyaceae). <em>Cryptogamie Algologie</em> 38 (4): 285–323. https://doi.org/10.7872/crya/v38.iss4.2017.285</p>
<p>Song, G.F., Jiang, Y.G. & Li, R. (2015) <em>Scytolyngbya timoleontis</em>, <em>gen. et sp. nov.</em> (Leptolyngbyaceae, Cyanobacteria): a novel false branching Cyanobacteria from China. <em>Phytotaxa</em> 224: 72–84. https://doi.org/10.11646/phytotaxa.224.1.5</p>
<p>Stamatakis, A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. <em>Bioinformatics </em>22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446</p>
<p>Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML web servers. <em>Systematic Biology</em> 57: 758–771. https://doi.org/10.1080/10635150802429642</p>
<p>Taton, A., Grubisic, S., Brambilla, E., De Wit, R. & Wilmotte, A. (2003) Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. <em>Applied Environmental Microbiology</em> 69: 5157–69. https://doi.org/10.1128/AEM.69.9.5157-5169.2003</p>
<p>Ulcay, S., Taşkin, E., Kurt, O. & Öztürk, M. (2015) Marine benthic cyanobacteria in Northern Cyprus (Eastern Mediterranean Sea). <em>Turkish Journal of Botany</em> 39: 173– 188. https://doi.org/10.3906/bot-1311-52</p>
<p>Vaz, M.G.M.V., Genuário, D.B., Andreote, A.P.D., Malone, C.F.S., Sant´Anna, C.L., Barbiero, L. & Fiore, M.F. (2015) <em>Pantanalinema</em> <em>gen. nov.</em> and <em>Alkalinema</em> <em>gen. nov.</em>: two novel pseudanabaenacean genera (Cyanobacteria) isolated from saline-alkaline lakes. <em>International Journal of Systematic and Evolutionary Microbiology</em> 65: 298–308. https://doi.org/10.1099/ijs.0.070110-0</p>
<p>Zammit, G., Billi, D. & Albertano, P. (2012) The subaerophytic cyanobacterium <em>Oculatella subterranea</em> (Oscillatoriales, Cyanophyceae) <em>gen. et sp. nov.</em>: a cytomorphological and molecular description. <em>European Journal of Phycology</em> 47: 341–354. https://doi.org/10.1080/09670262.2012.717106</p>
<p>Zhou, W., Ding, D., Yang, Q., Ahmad, M., Zhang, Y., Lin, X., Zhang, Y., Ling, J. & Dong, J. (2018) <em>Marileptolyngbya sina gen. nov., sp. nov. </em>and <em>Salileptolyngbya diazotrophicum gen. nov., sp. nov. </em>(Synechococcales, Cyanobacteria), species of cyanobacteria isolated from a marine ecosystem. <em>Phytotaxa</em> 383: 75–92. https://doi.org/10.11646/phytotaxa.383.1.4</p>
<p>Zubia, M., Turquet, J. & Golubic, S. (2016) Benthic cyanobacterial diversity of iles eparses [Scattered islands] in the Mozambique channel. <em>Acta Oecologica</em> 72: 21–32. https://doi.org/10.1016/j.actao.2015.09.004</p>
<p>Zubia, M., Vieira, C., Palinska, K.A., Roué, M., Gaertner, J.-C., Zloch, I. & Golubic, S. (2019) Benthic cyanobacteria on coral reefs of Moorea Island (French Polynesia): diversity response to habitat quality. <em>Hydrobiologia</em> 843: 61–78. https://doi.org/10.1007/s10750-019-04029-8</p>
<p>Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. <em>Nucleic Acids Research</em> 31: 3406–3415. https://doi.org/10.1093/nar/gkg595</p>