Abstract
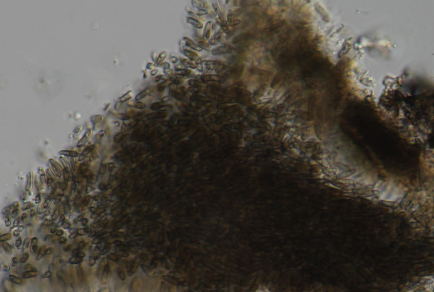
A saprobic fungal specimen was collected from dead wood in Mianyang City, Sichuan Province, during a survey on fungal diversity. Preliminary BLAST analysis of the internal transcribed spacer (ITS) region indicated that the isolate belongs to the genus Pseudophaeocytostroma (Diaporthaceae). To determine its phylogenetic position, multilocus phylogenetic analyses were performed using sequences from the ITS, the large subunit ribosomal RNA (LSU), and the translation elongation factor 1-alpha (tef1-α) regions. The resultant phylogeny revealed that the isolate forms an independent clade that is closely related to “Phaeocytostroma sacchari” and Pseudophaeocytostroma bambusicola. Morphologically, our collection differs from its closest relative by the size of conidia and the abundance of paraphyses. Single-gene phylogenies revealed topological incongruence among loci in Diaporthaceae, indicating the need for caution when delimiting new species and generic placements. This underscores the importance of incorporating additional molecular markers or genomic data in future taxonomic revisions. Based on combined molecular evidence and morphological characteristics, we hereby introduce Phaeocytostroma saprophyticum as a novel species.
References
- Aguiar, F.M., Lanza, F.E., Costa, R.V., Silva, D.D., Lana, U.G.P., Guimarães, E.A., Gomes, G.R. & Cota, L.V. (2016) First report of Phaeocytostroma ambiguum causing Maize Stalk Rot in Brazil. Plant Disease 100 (12): 2528–2528. https://doi.org/10.1094/PDIS-04-16-0533-PDN
- Barr, M.E. (1978) The Diaporthales in North America: with emphasis on Gnomonia and its segregates. Mycologia Memoir 7: 1–232.
- Bobev, S.G., Van, P.K. & Maes, M. (2016) First report of Phaeocytostroma ambiguum causing root and stem base rot on maize in Bulgaria. Plant Disease 100 (6): 1237–1237. https://doi.org/10.1094/PDIS-11-15-1327-PDN
- Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) trimAl: A Tool for automate alignment trimming in large-scale phylogenetic Analyses. Bioinformatics 25 (15): 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
- Carabez, J.S., Ascencio, S.O. & Pedraza, J.T. (2014) First report of stalk rot disease of sugarcane caused by Phaeocytostroma sacchari in Mexico. Plant Disease 98 (3): 420–420. https://doi.org/10.1094/PDIS-05-13-0563-PDN
- Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., Liu, Y., Feng, J., Chen, H., He, Y. & Xia, R. (2023) TBtools-II: A "one for all, all for one" bioinformatics platform for biological big-data mining. Molecular Plant 16: 1733–1742.
- Crous, P.W., Slippers, B., Wingfield, M.J., Rheeder, J., Marasas, W.F.O., Philips, A.J.L., Alves, A., Burgess, T., Barber, P. & Groenewald, J.Z. (2006) Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. https://doi.org/10.3114/sim.55.1.235
- de la Riva, A., García-Carneros, A.B. & Molibnero-Ruiz, L. (2020) First report of stalk rot of maize caused by Phaeocytostroma ambiguum in the Iberian Peninsula. Plant Disease 104 (4): 1255–1255. https://doi.org/10.1094/PDIS-10-19-2185-PDN
- Glez-Pena1, D., Go mez-Blanco, D., Reboiro-Jato, M., Fdez-Riverola, F. & Posada, D.A (2010) ALTER: program-oriented aonversion of DNA and protein alignments. Nucleic Acids Research 38 (suppl 2): W14–W18. https://doi.org/10.1093/nar/gkq321
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor andanalysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41 (41): 95–98.
- Hopple, J.S. & Vilgalys, R. (1999) Phylogenetic relationships in the mushroom Genus Coprinus and dark-Spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Molecular Phylogenetics and Evolution 13 (1): 1–19. https://doi.org/10.1006/mpev.1999.0634
- Hyde, K.D., Noorabadi, M.T., Thiyagaraja, V., He, M.Q., Johnston, P.R., Wijesinghe, S.N., Armand, A., Biketova, A.Y., Chethana, K.W.T., Erdoğdu, M., Ge, Z.W., Groenewald, J.Z., Hongsanan, S., Kušan, I., Leontyev, D.V., Li, D.W., Lin, C.G., Liu, N.G., Maharachchikumbura, S.S.N., Matočec, N., May, T.W., McKenzie, E.H.C., Mešić, A., Perera, R.H., Phukhamsakda, C., Piątek, M., Samarakoon, M.C., Selcuk, F., Senanayake, I.C., Tanney, J.B., Tian, Q., Vizzini, A., Wanasinghe, D.N., Wannasawang, N., Wijayawardene, N.N., Zhao, R.L., Abdel-Wahab, M.A., Abdollahzadeh, J., Abeywickrama, P.D., Abhinav, Absalan, S., Acharya, K., Afshari, N., Afshan, N.S., Afzalinia, S., Ahmadpour, S.A., Akulov, O., Alizadeh, A., Alizadeh, M., Al-Sadi, A.M., Alves, A., Alves, V.C.S., Alves-Silva, G., Antonín, V., Aouali, S., Aptroot, A., Apurillo, C.C.S., Arias, R.M., Asgari, B., Asghari, R., Assis, D.M.A., Assyov, B., Atienza, V., Aumentado, H.D.R., Avasthi, S., Azevedo, E., Bakhshi, M., Bao, D.F., Baral, H.O., Barata, M., Barbosa, K.D., Barbosa, R.N., Barbosa, F.R., Baroncelli, R., Barreto, G.G., Baschien, C., Bennett, R.M., Bera, I., Bezerra, J.D.P., Bhunjun, C.S., Bianchinotti, M.V., Błaszkowski, J., Boekhout, T., Bonito, G.M., Boonmee, S., Boonyuen, N., Bortnikov, F.M., Bregant, C., Bundhun, D., Burgaud, G., Buyck, B., Caeiro, M.F., Cabarroi-Hernández, M., Cai, M.F., Cai, L., Calabon, M.S., Calaça, F.J.S., Callalli, M., Câmara, M.P.S., Cano-Lira, J., Cao, B., Carlavilla, J.R., Carvalho, A., Carvalho, T.G., Castañeda-Ruiz, R.F., Catania, M.D.V., Cazabonne, J., Cedeño-Sanchez, M., Chaharmiri-Dokhaharani, S., Chaiwan, N., Chakraborty, N., Cheewankoon, R., Chen, C., Chen, J., Chen, Q., Chen, Y.P., Chinaglia, S., Coelho-Nascimento, C.C., Coleine, C., CostaRezende, D.H., Cortés-Pérez, A., Crouch., J.A., Crous, P.W., Cruz, R.H.S.F., Czachura, P., Damm, U., Darmostuk, V., Daroodi, Z., Das, K., Das, K., Davoodian, N., Davydov, E.A., da Silva, G.A., da Silva, I.R., da Silva, R.M.F., da Silva Santos, A.C., Dai, D.Q., Dai, Y.C., de Groot, Michiel, D., De Kesel, A., De Lange, R., de Medeiros, E.V., de Souza, C.F.A., de Souza, F.A., dela Cruz, T.E.E., Decock, C., Delgado, G., Denchev, C.M., Denchev, T.T., Deng, Y.L., Dentinger, B.T.M., Devadatha, B., Dianese, J.C., Dima, B., Doilom, M., Dissanayake, A.J., Dissanayake, D.M.L.S., Dissanayake, L.S., Diniz, A.G., Dolatabadi, S., Dong, J.H., Dong, W., Dong, Z.Y., Drechsler-Santos, E.R., Druzhinina, I.S., Du, T.Y., Dubey, M.K., Dutta, A.K., Elliott, T.F., Elshahed, M.S., Egidi, E., Eisvand, P., Fan, L., Fan, X., Fan, X.L., Fedosova, A.G., Ferro, L.O., Fiuza, P.O., Flakus, A.W., Fonseca, E.O., Fryar, S.C., Gabaldón, T., Gajanayake, A.J., Gannibal, P.B., Gao, F., GarcíaSánchez, D., García-Sandoval, R., Garrido-Benavent, I., Garzoli, L., Gasca-Pineda, J., Gautam, A.K., Gené, J., Ghobad-Nejhad, M., Ghosh, A., Giachini, A.J., Gibertoni, T.B., Gentekaki, E., Gmoshinskiy, V.I., GóesNeto, A., Gomdola, D., Gorjón, S.P., Goto, B.T., Granados-Montero, M.M., Griffith, G.W., Groenewald, M., Grossart, H.-P., Gu, Z.R., Gueidan, C., Gunarathne, A., Gunaseelan, S., Guo, S.L., Gusmão, L.F.P., Gutierrez, A.C., Guzmán-Dávalos, L., Haelewaters, D., Haituk, H., Halling, R.E., He, S.C., Heredia, G., HernándezRestrepo, M., Hosoya, T., Hoog, S.D., Horak, E., Hou, C.L., Houbraken, J., Htet, Z.H., Huang, S.K., Huang, W.J., Hurdeal, V.G., Hustad, V.P., Inácio, C.A., Janik, P., Jayalal, R.G.U., Jayasiri, S.C., Jayawardena, R.S., Jeewon, R., Jerônimo, G.H., Jin, J., Jones, E.B.G., Joshi, Y., Jurjević, Ž., Justo, A., Kakishima, M., Kaliyaperumal, M., Kang, G.P., Kang, J.C., Karimi, O., Karunarathna, S.C., Karpov, S.A., Kezo, K., Khalid, A.N., Khan, M.K., Khuna, S., Khyaju, S., Kirchmair, M., Klawonn, I., Kraisitudomsook, N., Kukwa, M., Kularathnage, N.D., Kumar, S., Lachance, M.A., Lado, C., Latha, K.P.D., Lee, H.B., Leonardi, M., Lestari, A.S., Li, C., Li, H. Li, J., Li, Q., Li, Y., Li, Y.C., Li, Y.X., Liao, C.F., Lima, J.L.R., Lima, J.M.S., Lima, N.B., Lin, L., Linaldeddu, B.T., Linn, M.M., Liu, F., Liu, J.K., Liu, J.W., Liu, S., Liu, S.L., Liu, X.F., Liu, X.Y., Longcore, J.E., Luangharn, T., Luangsa-ard, J.J., Lu, L., Lu, Y.Z., Lumbsch, H.T., Luo, L., Luo, M., Luo, Z.L., Ma, J., Madagammana, A.D., Madhushan, A., Madrid, H., Magurno, F., Magyar, D., Mahadevakumar, S., Malosso, E., Malysh, J.M., Mamarabadi, M., Manawasinghe, I.S., Manfrino, R.G., Manimohan, P., Mao, N., Mapook, A., Marchese, P., Marasinghe, D.S., Mardones, M., Marin-Felix, Y., Masigol, H., Mehrabi, M., MehrabiKoushki, M., Meiras-Ottoni, A., de Melo, R.F.R., Mendes-Alvarenga, R.L., Mendieta, S., Meng, Q.F., Menkis, A., Menolli, Jr. N., Mikšík, M., Miller, S.L., Moncada, B., Moncalvo, J.M., Monteiro, J.S., Monteiro, M., Mora-Montes, H.M., Moroz, E.L., Moura, J.C., Muhammad, U., Mukhopadhyay, S., Nagy, G.L., Najamul Sehar, A., Najafiniya, M., Nanayakkara, C.M., Naseer, A., Nascimento, E.C.R., Nascimento, S.S., Neuhauser, S., Neves, M.A., Niazi, A.R., Nie, Y., Nilsson, R.H., Nogueira, P.T.S., Novozhilov, Y.K., Noordeloos, M., Norphanphoun, C., Nuñez Otaño, N., O’Donnell, R.P., Oehl, F., Oliveira, J.A., Oliveira Junior, I., Oliveira, N.V.L., Oliveira P.H.F., Orihara, T., Oset, M., Pang, K.L., Papp, V., Pathirana, L.S., Peintner, U., Pem, D., Pereira, O.L., Pérez-Moreno, J., Pérez-Ortega, S., Péter, G., Pires-Zottarelli, C.L.A., Phonemany, M., Phongeun, S., Pošta, A., Prazeres, J.F.S.A., Quan, Y., Quandt, C.A., Queiroz, M.B., Radek, R., Rahnama, K., Raj, K.N.A., Rajeshkumar, K.C., Rajwar, S., Ralaiveloarisoa, A.B., Rämä, T., Ramírez-Cruz, V., Rambold, G., Rathnayaka, A.R., Raza, M., Ren, G.C., Rinaldi, A.C., Rivas-Ferreiro, M., Robledo, G.L., Ronikier, A., Rossi, W., Rusevska, K., Ryberg, M., Safi, A., Salimi, F., Salvador-Montoya, C.A., Samant, B., Samaradiwakara, N.P., Sánchez-Castro, I., Sandoval-Denis, M., Santiago, A.L.C.M.A., Santos, A.C.D.S., Santos, L.A., dos Sarma, V.V., Sarwar, S. Savchenko, A., Savchenko, K., Saxena, R.K., Schoutteten, N., Selbmann, L., Ševčíková, H., Sharma, A., Shen, H.W., Shen, Y.M., Shu, Y.X., Silva, H.F., Silva-Filho, A.G.S., Silva, V.S.H., Simmons, D.R., Singh, R., Sir, E.B., Sohrabi, M., Souza, F.A., Souza-Motta, C.M., Sriindrasutdhi, V., Sruthi, O.P., Stadler, M., Stemler, J., Stephenson, S.L., Stoyneva-Gaertner, M.P., Strassert, J.F.H., Stryjak-Bogacka, M., Su, H., Sun, Y.R., Svantesson, S., Sysouphanthong, P., Takamatsu, S., Tan, T.H., Tanaka, K., Tang, C., Tang, X., Taylor, J.E., Taylor, P.W.J., Tennakoon, D.S., Thakshila, S.A.D., Thambugala, K.M., Thamodini, G.K., Thilanga, D., Thines, M., Tiago, P.V., Tian, X.G., Tian, W.H., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Tomšovský, M., Torruella, G., Tsurykau, A., Udayanga, D., Ulukapı, M., Untereiner, W.A., Usman, M., Uzunov, B.A., Vadthanarat, S., Valenzuela, R., Van den Wyngaert, S., Van Vooren, N., Velez, P., Verma, R.K., Vieira, L.C., Vieira, W.A.S., Vinzelj, J.M., Tang, A.M.C., Walker, A., Walker, A.K., Wang, Q.M., Wang, Y., Wang, X.Y., Wang, Z.Y., Wannathes, N., Wartchow, F., Weerakoon, G., Wei, D.P., Wei, X., White, J.F., Wijesundara, D.S.A., Wisitrassameewong, K., Worobiec, G., Wu, H.X., Wu, N., Xiong, Y.R., Xu, B., Xu, J.P., Xu, R., Xu, R.F., Xu, R.J., Yadav, S., Yakovchenko, L.S., Yang, H.D., Yang, X., Yang, Y.H., Yang, Y., Yang, Y.Y., Yoshioka, R., Youssef Noha, H., Yu, F.M., Yu, Z.F., Yuan, L.L., Yuan, Q., Zabin, D.A., Zamora, J.C., Zapata, C.V., Zare, R., Zeng, M., Zeng, X.Y., Zhang, J.F., Zhang, J.Y., Zhang, S., Zhang, X.C., Zhao, C.L., Zhao, H., Zhao, Q., Zhao, H., Zhao, H.J., Zhou, H.M., Zhu, X.Y., Zmitrovich, I.V., Zucconi, L. & Zvyagina, E. (2024) The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 15 (1): 5146–6239. https://doi.org/10.5943/mycosphere/15/1/25
- Jiang, N., Voglmayr, H., Piao, C.G. & Li, Y. (2021) Two new species of Diaporthe (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. MycoKeys 85: 31. https://doi.org/10.3897/mycokeys.85.73107
- Jiang, N., Xue, H. & Li, Y. (2025) Novel genus and species of Diaporthostomataceae (Diaporthales) in China. IMA Fungus 16: e145422. https://doi.org/10.3897/imafungus.16.145422
- Katoh, K. (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research 30 (14): 3059–3066. https://doi.org/10.1093/nar/gkf436
- Kruger, W. (1965) Phaeocytostroma ambiguum (Mont.) Petr. A parasitic fungus of maize in South Africa-research note. South African Journal of Agricultural Science 8 (2): 587–592.
- Lamprecht, S.C., Crous, P.W., Groenewald, J.Z., Tewoldemedhin, Y.T. & Marasas, W.F. (2011) Diaporthaceae associated with root and crown rot of maize. IMA Fungus 2: 13–24. https://doi.org/10.5598/imafungus.2011.02.01.03
- Larsson, A. (2014) AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30 (22): 3276–3278.
- Madhushan, A., Weerasingha, D.B., Ilyukhin, E., Taylor, P.W., Ratnayake, A.S., Liu, J.K. & Maharachchikumbura, S.S.N. (2025) From natural hosts to agricultural threats: The evolutionary journey of phytopathogenic fungi. Journal of Fungi 11 (1): 25. https://doi.org/10.3390/jof11010025
- Melo, J.A.D.S., Abreu, V.P.D., Teles, T.A.S. & Cunha, M.G.D. (2023) Emergence of Phaeocytostroma sacchari in sugarcane plantations in Brazil. Journal of Plant Pathology 105 (3): 1163–1163. https://doi.org/10.1007/s42161-023-01366-5
- Melo, J.A.D.S., Abreu, V.P.D., Teles, T.A.S. & Cunha, M.G.D. (2024) Rapid detection of Phaeocytostroma sacchari in sugarcane using conventional polymerase chain reaction. Pesquisa Agropecuária Tropical 54: e78370. https://doi.org/10.1007/s42161-023-01366-5
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 gateway computing environments workshop (GCE), pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Monkai, J., Phookamsak, R., Xu, S., Xu, J., Mortimer, P.E., Karunarathna, S.C., Suwannarach, N. & Lumyong, S. (2022) Pseudophaeocytostroma bambusicola gen. sp. Nov. (Diaporthaceae) from bamboo in Yunnan, P.R. China. Phytotaxa 571 (1): 39–51. https://doi.org/10.11646/phytotaxa.571.1.3
- Petrak, F. (1921) Mykologische Notizen. II. Annales Mycologici 19: 17–128.
- Rehner, S.A. & Buckley, E.A. (2005) Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97 (1): 84–98. https://doi.org/10.1080/15572536.2006.11832842
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, F., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
- Rossman, A.Y., Farr, D.F. & Castlebury, L.A. (2007) A review of the phylogeny and biology of the Diaporthales. Mycoscience 48 (3): 135–144. https://doi.org/10.1007/s10267-007-0347-7
- Senanayake, I.C., Crous, P.W., Groenewald, J.Z., Maharachchikumbura, S.S.N., Jeewon, R., Phillips, A.J.L., Bhat, D.J., Perera, R.H., Li, Q.R., Li, W.J., Tangthirasunun, N., Norphanphoun, C., Karunarathna, S.C., Camporesi, E., Manawasighe, I.S., Al-Sadi, A.M. & Hyde, K.D. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. https://doi.org/10.1016/j.simyco.2017.07.003
- Senanayake, I.C., Jeewon, R., Chomnunti, P., Wanasinghe, D.N., Norphanphoun, C., Karunarathna, A., Pem, D., Perera, R.H., Camporesi, E., McKenzie, E.H.C. & Hyde, K.D. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93: 241–443. https://doi.org/10.1007/s13225-018-0410-z
- Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 11 (1): 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
- Smith, H., Wingfield, M.J., Coutinho, T.A. & Crous, P.W. (1996) Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62 (2): 86–88. https://doi.org/10.1016/S0254-6299(15)30596-2
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9): 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57 (5): 758–771. https://doi.org/10.1080/10635150802429642
- Sutton, B.C. (1964) Coelomycetes III. Annellolacinia gen. nov., Aristastoma, Phaeocytostroma, Seimatosporium, etc. CABI Databases 42.
- Sutton, B.C. (1981) The Coelomycetes. Fungi imperfecti with pycnidia, acervuli and stromata. Commonwealth Mycological Institute, Kew, England, 704 pp.
- Thambugala, K.M. & Hyde, K.D. (2018) Additions to the genus Massariothea in Diaporthaceae. Mycological Progress 17 (10): 1139–1147. https://doi.org/10.1007/s11557-018-1426-1
- Toju, H., Tanabe, A.S., Yamamoto, S. & Sato, H. (2012) High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PloS One 7 (7): e40863. https://doi.org/10.1371/journal.pone.0040863
- Van Niekerk, J.M., Verkley, G.J., Fourie, P.H., Wingfield, M.J. & Crous, P.W. (2004) Systematic reappraisal of Coniella and Pilidiella, with specific reference to species occurring on Eucalyptus and Vitis in South Africa. Mycological Research 108 (3): 283–303. https://doi.org/10.1017/S0953756204009268
- Viswanathan, R., Premachandran, M.N., Balamuralikrishnan, M. & Jothi, R. (2003) A new stalk rot disease of sugarcane caused by Phaeocytostroma sacchari in India. Sugar Tech 5: 61–64. https://doi.org/10.1007/BF02943766
- von Höhnel, F.X.R. (1917) System der Diaportheen. Borntraeger 35: 631–638.
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sánchez-García, M., Goto, B.T., Saxena, R.K., Erdoðdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., Błaszkowski, J., Boonyuen, N., da Silva, G.A., de Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Mešić, A., Monteiro, J.S., Oehl, F., Pawłowska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro, I., Tang, L.Z., Stadler, M., Yurkov, A. & Thines, M. (2022) Outline of Fungi and fungus-like taxa‒2021. Mycosphere 13: 53–453. https://doi.org/10.5943/mycosphere/13/1/2
- Xiang, C., Gao, F., Jakovlić, I., Lei, H., Hu, Y., Zhang, H., Zou, H., Wang, G. & Zhang, D. (2023) Using phyloSuite for molecular phylogeny and tree‐based analyses. iMeta 2 (1): e87.
- Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20 (1): 348–355. https://doi.org/10.1111/1755-0998.13096
- Zhang, Z.X., Shang, Y., Liu, Q.Y., Li, D.H., Yin, C.Z., Liu, X.Y., Tao, M.F., Jiang, Y., Wang, Y.X., Zhang, M.F., Jiang, Y., Wang, Y.X., Zhang, M.Y., Dong, Z.X., Yun, J.X., Xia, J.W., Wang, S., Li, Z., Luo, Z.L., Liu, X.Y. & Zhang, X.G. (2025) Deciphering the evolutionary and taxonomic complexity of Diaporthales (Sordariomycetes, Ascomycota) through integrated phylogenomic and divergence time estimation. Fungal Diversity: 1–125. https://doi.org/10.1007/s13225-025-00551-4
- Zhu, Y.Q., Ma, C.Y., Xue, H., Piao, C.G., Li, Y. & Jiang, N. (2023) Two new species of Diaporthe (Diaporthaceae, Diaporthales) in China. MycoKeys 95: 209. https://doi.org/10.3897/mycokeys.95.98969