Abstract
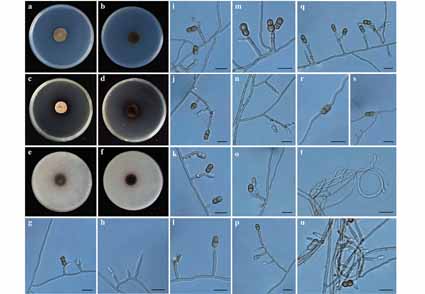
A new species of Verruconis, V. guizhouensis, isolated from the rhizosphere soil of Viola philippica is introduced. Morphological characteristics and phylogenetic analyses based on multi-locus datasets (SSU + ITS + LSU) support the establishment of the new species. Phylogenetically, V. guizhouensis formed a separated subclade which has a sister relationship to V. verruculosa, V. terricola and V. thailandica with 100% MLBS and 1.00 BYPP support values. Morphologically, V. guizhouensis is similar to its three sister species, with brown, 1-septate, constricted at the septum, verrucose conidia. However, it is clearly distinguished from these three species by the presence of intercalary conidia and ovoidal conidia.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N, McKenzie, E.H.C., Sarma, V.V., Lücking, R., Boonmee, S., Bhat, J.D., Liu, N.G., Tennakoon, D.S., Pem, D., Karunarathna, A., Jiang, S.H., Jones, G.E.B., Phillips, A.J.L., Manawasinghe, I.S., Tibpromma, S., Jayasiri, S.C., Sandamali, D., Jayawardena, R.S., Wijayawardene, N.N., Ekanayaka, A.H., Jeewon, R., Lu, Y.Z., Phukhamsakda, C., Dissanayake, A.J., Zeng, X.Y., Luo, Z.L., Tian, Q., Thambugala, K.M., Dai, D., Samarakoon, M.C., Chethana, K.W.T., Ertz, D., Doilom, M., Liu, J.K., Pérez-Ortega, S., Suija, A., Senwanna, C., Wijesinghe, S.N., Niranjan, M., Zhang, S.N., Ariyawansa, H.A., Jiang, H.B., Zhang, J.F., Norphanphoun, C., de Silva, N., Thiyagaraja, V., Zhang, H., Bezerra, J.D.P., Miranda-González, R., Aptroot, A., Kashiwadani, H., Harishchandra, D., Sérusiaux, E., Abeywickrama, P.D., Bao, D.F., Devadatha, B., Wu, H.X., Moon, K.H., Gueidan, C., Schumm, F., Bundhun, D., Mapook, A., Monkai, J., Bhunjun, C.S., Chomnunti, P., Suetrong, S., Chaiwan, N., Dayarathne, M.C., Yang, J., Rathnayaka, A.R., Xu, J.C., Zheng, J., Liu, G., Feng, Y. & Xie, N. (2020) Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105: 17–318. https://doi.org/10.1007/s13225-020-00462-6</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Huanraluek, N., Phukhamsakda, C., Senwanna, C., Hongsanan, S., Jayawardena, R.S., Bhat, D.J. & Hyde, K.D. (2019) Verruconis heveae, a novel species from Hevea brasiliensis in Thailand. Phytotaxa 403 (1): 47–54. https://doi.org/10.11646/phytotaxa.403.1.4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hyde, K.D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D.J., Jones, E.B.G., Liu, N.G., Abeywickrama, P.D., Mapook, A., Wei, D., Perera, R.H., Manawasinghe, I.S., Pem, D., Bundhun, D., Karunarathna, A., Ekanayaka, A.H., Bao, D.F., Li, J., Samarakoon, M.C., Chaiwan, N., Lin, C.G., Phutthacharoen, K., Zhang, S.N., Senanayake, I.C., Goonasekara, I.D., Thambugala, K.M., Phukhamsakda, C., Tennakoon, D.S. Jiang, H.B., Yang, J., Zeng, M., Huanraluek, N., Liu, J.K., Wijesinghe, S.N., Tian, Q., Tibpromma, S., Brahmanage, R.S., Boonmee, S., Huang, S.K., Thiyagaraja, V., Lu, Y.Z., Jayawardena, R.S., Dong, W., Yang, E.F., Singh, S.K., Singh, S.M., Rana, S., Lad, S.S., Anand, G., Devadatha, B., Niranjan, M., Sarma, V.V., Liimatainen, K., Aguirre-Hudson, B., Niskanen, T., Overall, A., Alvarenga, R.L.M., Gibertoni, T.B., Pfliegler, W.P., Horváth, E., Imre, A., Alves, A.L., da Silva Santos, A.C., Tiago, P.V., Bulgakov, T.S., Wanasinghe, D.N., Bahkali, A.H., Doilom, M., Elgorban, A.M., Maharachchikumbura, S.S.N., Rajeshkumar, K.C., Haelewaters, D., Mortimer, PE., Zhao, Q., Lumyong, S., Xu, J. & Sheng, J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–77. https://doi.org/10.1007/s13225-020-00439-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30 (4): 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, H., Brettell, L.E., Qiu, Z. & Singh, B.K. (2020) Microbiome-mediated stress resistance in plants. Trends in Plant Science 25 (8): 733–743. https://doi.org/10.1016/j.tplants.2020.03.014</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Mahmud, K., Missaoui, A., Lee, K., Ghimire, B., Presley, H.W. & Makaju, S. (2021) Rhizosphere microbiome manipulation for sustainable crop production. Current Plant Biology 27: 100210. https://doi.org/10.1016/j.cpb.2021.100210</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular biology and evolution 32 (1): 268–274. https://doi.org/10.1093/molbev/ msu300</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Qiao, M., Tian, W., Castañeda-Ruiz, R.F., Xu, J. & Yu, Z. (2019) Two new species of Verruconis from Hainan, China. MycoKeys 48: 41–53. https://doi.org/10.3897/mycokeys.48.32147</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ren, J., Jie, C.Y., Zhou, Q.X., Li, X.H., Hyde, K.D., Jiang, Y.L., Zhang, T.Y. & Wang, Y. (2013) Molecular and morphological data reveal two new species of Scolecobasidium. Mycoscience 54 (6): 420–425. https://doi.org/10.1016/j.myc.2013.01.007</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Samerpitak, K., der Linde, E.V., Choi, H.J., van den Ende, A.H.G.G., Machouart, M., Gueidan, C. & De Hoog, G.S. (2014) Taxonomy of Ochroconis, genus including opportunistic pathogens on humans and animals. Fungal Diversity 65: 89–126. https://doi.org/10.1007/s13225-013-0253-6</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Seyedmousavi, S., Samerpitak, K., Rijs, A.J.M.M., Melchers, W.J.G., Mouton, J.W., Verweij, P.E. & de Hoog, G.S. (2014) Antifungal susceptibility patterns of opportunistic fungi in the genera Verruconis and Ochroconis. Antimicrobial Agents and Chemotherapy 58 (6): 3285–3292. https://doi.org/10.1128/AAC.00002-14</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Shen, M., Zhang, J.Q., Zhao, L.L., Groenewald, J.Z., Crous, P.W. & Zhang, Y. (2020) Venturiales. Studies in Mycology 96: 185–308. https://doi.org/10.1016/j.simyco.2020.03.001</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30 (12): 2725–2729. https://doi.org/10.1093/molbev/mst197</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T., Tedersoo, L., Haelewaters, D., Rajeshkumar, K.C., Zhao, R.L., Aptroot, A., Leontyev, D.V., Saxena, R.K., Tokarev, Y.S., Dai, D.Q., Letcher, P.M., Stephenson, S.L., Ertz, D., Lumbsch, H.T., Kukwa, M., Issi, I.V., Madrid, H., Phillips, A.J.L., Selbmann, L., Pfliegler, W.P., Horváth, E., Bensch, K., Kirk, P.M., Kola?íková, K., Raja, H.A., Radek, R., Papp, V., Dima, V., Ma, J., Malosso, E., Takamatsu, S., Rambold, G., Gannibal, P.B., Triebel, D., Gautam, A.K., Avasthi, S., Suetrong, S., Timdal, E., Fryar, S.C., Delgado, G., Réblová, M., Doilom, M., Dolatabadi, S., Paw?owska, J.Z., Humber, R.A., Kodsueb, R., Sánchez-Castro, I., Goto, B.T., Silva, D.K.A., de Souza, F.A., Oehl, F., da Silva, G.A., Silva, I.R., B?aszkowski, J., Jobim, K., Maia, L.C., Barbosa, F.R., Fiuza, P.O., Divakar, P.K., Shenoy, B.D., Castañeda-Ruiz, R.F., Somrithipol, S., Lateef, A.A., Karunarathna, S.C., Tibpromma, S., Mortimer, P.E., Wanasinghe, D.N., Phookamsak, R., Xu, J., Wang, Y., Tian, F., Alvarado, P., Li, D.W., Kušan, I., Mato?ec, N., Maharachchikumbura, S.S.N., Papizadeh, M., Heredia, G., Wartchow, F., Bakhshi, M., Boehm, E., Youssef, N., Hustad, V.P., Lawrey, J.D., Santiago, A.L.C.M.A., Bezerra, J.D.P., Souza-Motta, C.M., Firmino, A.L., Tian, Q., Houbraken, J., Hongsanan, S., Tanaka, K., Dissanayake, A.J., Monteiro, J.S., Grossart, H.P., Suija, A., Weerakoon, G., Etayo, J., Tsurykau, A., Vázquez, V., Mungai, P., Damm, U., Li, Q.R., Zhang, H., Boonmee, S., Lu, Y.Z., Becerra, A.G., Kendrick, B., Brearley, F.Q., Motiej?naitë, J., Sharma, B., Khare, R., Gaikwad, S., Wijesundara, D.S.A., Tang, L.Z., He, M.Q., Flakus, A., Rodriguez-Flakus, P., Zhurbenko, M.P., McKenzie, E.H.C., Stadler, M., Bhat, D.J., Liu, J.K., Raza, M., Jeewon, R., Nassonova, E.S., Prieto, M., Jayalal, R.G.U., Erdoðdu, M., Yurkov, A., Schnittler, M., Shchepin, O.N., Novozhilov, Y.K., Silva-Filho, A.G.S., Liu, P., Cavender, J.C., Kang, Y., Mohammad, S., Zhang, L.F., Xu, R.F., Li, Y.M., Dayarathne, M.C., Ekanayaka, A.H., Wen, T.C., Deng, C.Y., Pereira, O.L., Navathe, S., Hawksworth, D.L., Fan, X.L., Dissanayake, L.S., Kuhnert, E., Grossart, H.P. & Thines, M. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11 (1): 1060–1456. https://dx.doi.org/10.5943/mycosphere/11/1/8</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yarita, K., Sano, A., Samerpitak, K., Kamei, K., de Hoog, G.S. & Nishimura, K. (2010) Ochroconis calidifluminalis, a sibling of the neurotropic pathogen O. gallopava, isolated from hot spring. Mycopathologia 170 (1): 21–30. https://doi.org/10.1007/s11046-010-9292-7</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, T.Y., Yu, Y., Zhang, M.Y., Cheng, J., Chen, Z.J., Zhang, J.Y. & Zhang, Y.X. (2018) Verruconis panacis sp. nov., an endophyte isolated from Panax notoginseng. International Journal of Systematic Evolutionary Microbiology 68 (8): 2499–2503. https://doi.org/10.1099/ijsem.0.002862</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, X., Wang, K.Y., Ren, P.P. & Jiang,Y.L. (2020) Ochroconis terricola sp. nov. from China. Mycotaxon 135 (1): 143–150. https://doi.org/10.5248/135.143</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, Z.Y., Shao, Q.Y., Li, X., Chen, W.H., Liang, J.D., Han, Y.F., Huang, J.Z. & Liang, Z.Q. (2021) Culturable fungi from urban soils in China I: description of 10 new taxa. Microbiology Spectrum 9: e00867–00821. https://doi.org/10.1128/Spectrum.00867-21</span></span></span>