Abstract
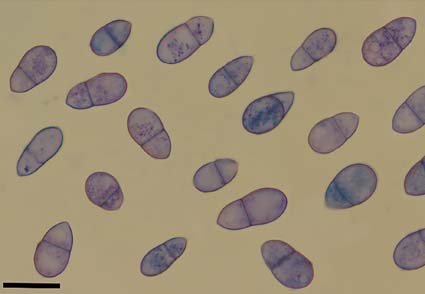
Arthrobotrys jindingensis and A. tongdianensis, two new species isolated from the freshwater sediment and terrestrial soil in Lanping county, Yunnan, China, are introduced based on morphology and multi-locus phylogenetic analysis (ITS, TEF and RPB2). A. jindingensis is characterized by its branched conidiophores with irregularly swollen nodular apex, pyriform or obovoid, and 1-septate conidia. A. tongdianensis is characterized by its branched conidiophores, and ellipsoid or obovate, 1-septate conidia. Phylogenetic analysis supports them as two new species within Arthrobotrys with high support. The descriptions and illustrations of these two species are provided in this paper.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Barron, G.L. (1979) Nematophagous fungi: a new Arthrobotrys with nonseptate conidia. Canadian Journal of Botany 57 (12): 1371–1373. https://doi.org/10.1139/b79-170</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Corda, A.K.J. (1839) Pracht-flora europaeischer Schimmelbildungen. G. Fleischer, 43 pp. http://dx.doi.org/10.1080/00222933909512494</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Drechsler, C. (1933) Morphological features of some more fungi that capture and kill nematode. Journal of the Washington Academy of Sciences 23: 267–270.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Drechsler, C. (1937) Some Hyphomycetes that Prey on Free-Living Terricolous Nematodes. Mycologia 29 (4): 447–552. https://doi.org/10.1080/00275514.1937.12017222</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Duddington, C.L. (1951) Two new predacious hyphomycetes. Transactions of the British Mycological Society 34 (4): 598–603. https://doi.org/10.1016/S0007-1536(51)80044-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Felsenstein, J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Fresenius, G. (1852) Bertrage Zur Mvkologie. Heft, 1–2.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hall, B.G. (2007) Phylogenetic Trees Made Easy: A How-to Manual; Massachusetts: Sinauer Associates Sunderland: Sunderland, MA, USA.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. https://doi.org/ 10.12691/ajmr-3-2-1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hao, Y.E., Luo, J. & Zhang, K.Q. (2004) A New Aquatic Nematode-trapping Hyphomycete. Mycotaxon 89 (2): 235–239.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30 (4): 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Li, J., Liu, Y., Zhu, H. & Zhang, K.Q. (2016) Phylogenic analysis of adhesion related genes Mad1 revealed a positive selection for the evolution of trapping devices of nematode-trapping fungi. Scientific Reports 6: 22609. https://doi.org/10.1038/srep22609.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Lin, H.B., Li, Y.P., Duan, J.X., Xia, F.Y., Xiong, H.J., Wang, J.B., Yang, X.Y. & Xiao, W. (2016) Screening of Nematode-trapping Fungi for Biocontrol Intestinal Parasitic Nematode of Yunnan Snub-nosed Monkey. Sichuan Journal of Zoology 35 (1): 31–37.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liou, G.Y. & Tzean, S.S. (1997) Phylogeny of the genus Arthrobotrys and allied nematode-trapping fungi based on rDNA sequences. Mycologia 89: 876–884. https://doi.org/10.1080/00275514.1997.12026858.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, S.R., Su, H.Y., Su, X.J., Zhang, F., Liao, G.H. & Yang, X.Y. (2014) Arthrobotrys xiangyunensis, a novel nematode-trapping taxon from a hot-spring in Yunnan Province, China. Phytotaxa 174 (2): 89–96. http://dx.doi.org/10.11646/phytotaxa.174.2.3</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular biology and evolution 16 (12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nguyen, L.T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular biology and evolution 32 (1): 268–274. https://doi.org/10.1093/molbev/msu300</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">O’Donnell, K., Kistler, H.C., Cigelnik, E. & Ploetz, R.C. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences 95 (5): 2044–2049. https://doi.org/10.1073/pnas.95.5.2044</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Pfister, D.H. (1997) Castor, Pollux and life histories of fungi. Mycologia 89: 1–23. https://doi.org/10.1080/00275514.1997.12026750</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Posada, D. (2008) jModelTest: phylogenetic model averaging. Molecular biology and evolution 25 (7): 1253–1256. https://doi.org/10.1093/molbev/msn083</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Quijada, L., Baral, H.O., Beltrán-Tejera, E. & Pfister, D.H. (2020) Orbilia jesu-laurae (Ascomycota, Orbiliomycetes), a new species of neo-tropical nematode-trapping fungus from Puerto Rico, supported by morphology and molecular phylogenetics. Willdenowia 50: 241–251. https://doi.org/10.3372/wi.50.50210</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rambaut, A. (2021) FigTree v1. 3.1. 2010. Available Online: http://tree. bio.ed.ac.uk/software/figtree/ (accessed 5 March 2021).</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rannala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10.1007/BF02338839</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rezakhani, F., Khodaparast, S.A., Masigol, H., Roja-Jimenez, K., Grossart, H.P. & Bakhshi, M. (2019) A preliminary report of aquatic hyphomycetes isolated from Anzali lagoon (Gilan province, North of Iran). Rostaniha 20: 123–143. https://doi.org/10.22092/BOTANY.2019.126701.1161.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Schenck, S., Kendrick, W.B. & Pramer, D. (1977) A new nematode-trapping hyphomycete and a reevaluation of Dactylaria and Arthrobotrys. Canadian Journal of Botany 55 (8): 977–985. https://doi.org/10.1139/b77-115</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Scholler, M., Hagedorn, G. & Rubner, A. (1999) A reevaluation of predatory orbiliaceous fungi: A new generic concept. Sydowia 51 (1): 89–113.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Su, H.Y., Liu, S.R., Li, Y.X. & Cao, Y.H. (2011) Arthrobotrys latispora, a new nematode-trapping fungus from southwest China. Mycotaxon 117 (3): 29–36. https://dx.doi.org/10.5248/117.29</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Swe, A., Jeewon, R. & Hyde, K.D. (2008) Nematode-Trapping fungi from mangrove habitats. Cryptogamie Mycologie 29 (4): 333–354. https://doi.org/10.1007/s10531-008-9553-7</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Swe, A., Jeewon, R., Pointing, S.B. & Hyde, K.D. (2008) Taxonomy and molecular phylogeny of Arthrobotrys mangrovispora, a new marine nematode-trapping fungal species. Botanica Marina 51: 331–338. https://doi.org/10.1515/BOT.2008.043</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Gronewald, J.Z., Cardinali, G. & Houbraken, J. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. https://doi.org/10.1016/j.simyco.2018.05.001</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wang, X., Li, G.H., Zou, C.G., Ji, X.L., Liu, T., Zhao, P.J., Liang, L.M., Xu, J.P., An, Z.Q., Zheng, X., Qin, Y.K., Tian, M.Q., Xu, Y.Y., Ma, Y.C., Yu, Z.F., Huang, X.W., Liu, S.Q., Niu, X.M., Yang, J.K., Ying Huang, Y. & Zhang, K.Q. (2014) Bacteria can mobilize nematode-trapping fungi to kill nematodes. Nature communications 5 (1): 1–9. https://doi.org/ 10.1038/ncomms6776</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18 (1): 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wu, H.Y., Kim, D.G., Ryu, Y.H. & Zhou, X.B. (2012) Arthrobotrys koreensis, a new nematode-trapping species from Korea. Sydowia 64: 129–136.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yang, E., Xu, L.L., Yang, Y., Zhang, X.Y., Xiang, M.C., Wang, C.S., An, Z.Q. & Liu, X.Z. (2012) Origin and evolution of carnivorism in the Ascomycota (fungi). PNAS 109: 10960–10965. https://doi.org/10.1073/pnas.1120915109.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yang, Y., Yang, E., An, Z. & Liu, X.Z. (2007) Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. PNAS 104: 8379–8384. https://doi.org/10.1073/pnas.0702770104.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, F., Boonmee, S., Bhat, J.D., Xiao, W. & Yang, X.Y. (2022) New Arthrobotrys Nematode-Trapping Species (Orbiliaceae) from Terrestrial Soils and Freshwater Sediments in China. Journal of Fungi 8 (7): 671. https://doi.org/10.3390/jof8070671</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, F., Zhou, X.J., Monkai, J., Li, F.T., Liu, S.R., Yang, X.Y., Xiao, W. & Hyde, K.D. (2020) Two new species of nematode-trapping fungi (Dactylellina, Orbiliaceae) from burned forest in Yunnan, China. Phytotaxa 452 (1): 65–74. https://doi.org/10.11646/phytotaxa.452.1.6</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, K.Q. & Hyde, K.D. (2014) Nematode-trapping fungi. Springer Science & Business, Berlin.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, K.Q. & Mo, M.H. (2006) Flora fungorum sinicorum (Vol. 33): Arthrobotrys et gengra cetera cognata. Science Press, Beijing.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, K.Q. (1997) Taxonomic Studies on Nematodephagous Hyphomycetes and Feasibility of Applying Some Molecular Biological Analysis. China Agricultural University.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhaxybayeva, O. & Gogarten, J.P. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. Genomics 3: 1–15. https://doi.org/10.1186/1471-2164-3-4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhu, M.L., Miao, Z.Q., Zhang, K.Q., Liu, X.Z. & Li, T.F. (2000a) Systematic studies of nematophagous fungi using RELP analysis of the PCR-AMPLIFIED ITS region of ribosomal DNA. Mycosystema 19 (2): 175–180. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhu, M.L., Miao, Z.Q., Zhang, K.Q., Liu, X.Z. & Li, T.F. (2000b) Phylogenetic Relationships between Monacrosporium Eudermatum and Arthrobotrys Musiformis revealed by RAPD. Mycosystema 19 (2): 342–347.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zopf, W.F. (1888) Zur Kenntniss der Infections-Krankheiten niederer Thiere und Pflanzen. Acad. Nat 52: 314–376.</span></span></span>