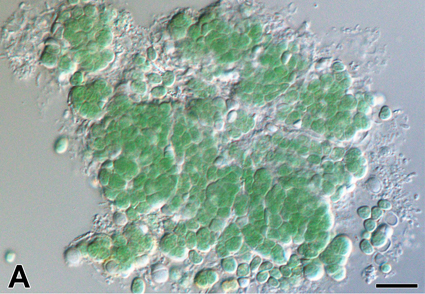
Abstract
A new coccoid cyanobacterium Aliterella vladivostokensis sp. nov. was described from an urban aerophytic habitat in a temperate monsoon climate (Vladivostok, Russia) using a polyphasic approach. Phylogenetic analyses based on the 16S rRNA gene sequences confirmed that our isolate was a member of the Aliterella genus clade. Aliterella species are hardly distinguishable from each other morphologically and were described from highly contrasting natural and artificial environments with only a few records from several continents. Despite high similarity of morphometric data for A. vladivostokensis and A. antarctica cells and a compensatory base change in the D1–D1′ helix shared by these species; high percent of dissimilarity (11.6±1.3) between their 16S–23S internal transcribed spacer sequences with at least 5 autapomorphic mutations in the D1–D1′ and Box-B helices, and distinct folding patterns of the Box-B helix allowed us to erect a new species.
References
Abdullin, Sh.R., Nikulin, V.Yu., Nikulin, A.Yu., Manyakhin, A.Yu., Bagmet, V.B., Suprun, A.R. & Gontcharov, A.A. (2021) Roholtiella mixta sp. nov. (Nostocales, Cyanobacteria): morphology, molecular phylogeny, and carotenoid content. Phycologia 60: 73–82. https://doi.org/10.1080/00318884.2020.1852846
Alvarenga, D.O., Fiore, M.F. & Varani, A.M. (2017) A Metagenomic Approach to Cyanobacterial Genomics. Frontiers in Microbiology 8: 809. https://doi.org/10.3389/fmicb.2017.00809
Becerra-Absalón, I., Johansen, J.R., Osorio-Santos, K. & Montejano, G. (2020) Two new Oculatella (Oculatellaceae, Cyanobacteria) species in soil crusts from tropical semi-arid uplands of México. Fottea 20: 160–170. https://doi.org/10.5507/fot.2020.010
Bohunická, M., Pietrasiak, N., Johansen, J.R., Gómez, E.B., Hauer, T., Gaysina, L.A. & Lukešová, A. Roholtiella, gen. nov. (Nostocales, Cyanobacteria)—a tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa 197: 84–103. https://doi.org/10.11646/phytotaxa.197.2.2
Bonfield, J.K., Smith, K.F & Staden, R. (1995) A new DNA sequence assembly program. Nucleic Acids Research 23: 4992–4999. https://doi.org/10.1093/nar/23.24.4992
Cellamare, M., Duval, C., Drelin, Y., Djediat, C., Touibi, N., Agogué, H., Leboulanger, C., Ader, M. & Bernard, C. (2018) Characterization of phototrophic microorganisms and description of new cyanobacteria isolated from the saline-alkaline crater-lake Dziani Dzaha (Mayotte, Indian Ocean). FEMS microbiology ecology 94. https://doi.org/10.1093/femsec/fiy108
Cirés, S. & Ballot, A. (2016) A review of the phylogeny, ecology and toxin production of bloom-forming Aphanizomenon spp. and related species within the Nostocales (cyanobacteria). Harmful Algae 54: 21–43. https://doi.org/10.1016/j.hal.2015.09.007
Coelho, C., Mesquita, N., Costa, I., Soares, F., Trovao, J., Freitas, H., Portugal, A. & Tiago, I. (2021) Bacterial and archaeal structural diversity in several biodeterioration patterns on the limestone walls of the Old Cathedral of Coimbra. Microorganisms 9: 709. https://doi.org/10.3390/microorganisms9040709
Czerwik-Marcinkowska, J. & Massalski, A. (2018) Diversity of cyanobacteria on limestone caves. In: Tiwari, A. (Ed.) Cyanobacteria. IntechOpen, London, pp. 137–164. https://doi.org/10.5772/intechopen.79750
Dadheech, P.K., Abed, R.M.M., Mahmoud, H., Mohan, M.K. & Krienitz, L. (2012) Polyphasic characterization of cyanobacteria isolated from desert crusts, and the description of Desertifilum tharense gen. et sp. nov. (Oscillatoriales). Phycologia 51: 260–270. https://doi.org/10.2216/09-51.1
Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. https://doi.org/10.1038/nmeth.2109
Darty, K., Denise, A. & Ponty, Y. (2009) VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975. https://doi.org/10.1093/bioinformatics/btp250
Duan, Y.L., Wu, F.S., He, D.P., Gu, J.D., Feng, H.Y., Chen, T., Liu, G.X. & Wang, W.F. (2021) Bacterial and fungal communities in the sandstone biofilms of two famous Buddhist grottoes in China. International Biodeterioration & Biodegradation 163: 105267. https://doi.org/10.1016/j.ibiod.2021.105267
Echt, C.S., Erdahl, L.A. & McCoy, T.J. (1992) Genetic segregation of random amplified polymorphic DNA in diploid cultivated alfalfa. Genome 35: 84–87. https://doi.org/10.1139/g92-014
Erwin, P.M. & Thacker, R.W. (2008) Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Molecular Ecology 17: 2937–2947. https://doi.org/10.1111/j.1365-294X.2008.03808.x
Galtier, N., Gouy, M. & Gautier, C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Computer applications in the biosciences: CABIOS 12: 543–548. https://doi.org/10.1093/bioinformatics/12.6.543
González-Gómez, W.S., Quintana, P., Gómez-Cornelio, S., García-Solis, C., Sierra-Fernandez, A., Ortega-Morales, O. & De la Rosa-García, S.C. (2018) Calcium oxalates in biofilms on limestone walls of Maya buildings in Chichén Itzá, Mexico. Environmental Earth Sciences 77: 230. https://doi.org/10.1007/s12665-018-7406-6
Harke, M.J., Steffen, M.M., Gobler, C.J., Otten, T.G., Wilhelm, S.W., Wood, S.A. & Paerl, H.W. (2016) A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 54: 4–20. https://doi.org/10.1016/j.hal.2015.12.007
Hauer, T., Mühlsteinová, R., Bohunická, M., Kaštovský, J. & Mareš, J. (2015) Diversity of cyanobacteria on rock surfaces. Biodiversity and Conservation 24: 759–779. https://doi.org/10.1007/s10531-015-0890-z
Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Iteman, I., Rippka, R., de Marsac, N.T. & Herdman, M. (2000) Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. Microbiology 146: 1275–1286. https://doi.org/10.1099/00221287-146-6-1275
Johansen, J.R., Bohunická, M., Lukešová, A., Hr?ková, K., Vaccarino, M.A. & Chesarino, N.M. (2014) Morphological and molecular characterization within 26 strains of the genus Cylindrospermum (Nostocaceae, Cyanobacteria), with description of three new species. Journal of Phycology 50: 187–202. https://doi.org/10.1111/jpy.12150
Jung, P., Mikhailyuk, T., Emrich, D., Baumann, K., Dultz, S. & Büdel, B. (2020) Shifting Boundaries: Ecological and Geographical Range extension Based on Three New Species in the Cyanobacterial Genera Cyanocohniella, Oculatella, and, Aliterella. Journal of Phycology 56: 1216–1231. https://doi.org/10.1111/jpy.13025
Kiselev, K.V., Dubrovina, A.S. & Tyunin, A.P. (2015) The methylation status of plant genomic DNA influences PCR efficiency. Journal of Plant Physiology 175: 59–67. https://doi.org/10.1016/j.jplph.2014.10.017
Komárek, J., Johansen, J.R., Šmarda, J. & Strunecký, O. (2020) Phylogeny and taxonomy of Synechococcus-like cyanobacteria. Fottea 20: 171–191. https://doi.org/10.5507/fot.2020.006
Komárek, J., Ková?ik, ?., Elster, J. & Komárek, O. (2012) Cyanobacterial diversity of Petuniabukta, Billefjorden, central Spitsbergen. Polish Polar Research 33: 347–368. https://doi.org/10.2478/V10183-012-0024-1
Köppen, W. (1936) Das geographische System der Klimate. In: Köppen, W. & Geiger, R. (Eds.) Handbuch der Klimatologie Bd. 1: Teil C. Verlag von Gebrüder Borntraeger, Berlin, pp. 1–44.
Kozlov, A.M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. https://doi.org/10.1093/bioinformatics/btz305
Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
Lane, D.J. (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E. & Goodfellow, M. (Eds.) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, pp. 115–175.
Lee, N.-J., Seo, Y., Ki, J.-S. & Lee, O.-M. (2020) Morphology and molecular description of Wilmottia koreana sp. nov. (Oscillatoriales, Cyanobacteria) isolated from the Republic of Korea. Phytotaxa 447: 237–251. https://doi.org/10.11646/phytotaxa.447.4.2
Li, Y., Cha, Q.-Q., Dang, Y.-R., Chen, X.-L., Wang, M., McMinn, A., Espina, G., Zhang, Y.-Z., Blamey, J.M. & Qin, Q.-L. (2019) Reconstruction of the Functional Ecosystem in the High Light, Low Temperature Union Glacier Region, Antarctica. Frontiers in Microbiology 10: 2408. https://doi.org/10.3389/fmicb.2019.02408
McFadden, G.I. & Melkonian, M. (1986) Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia 25: 551–557. https://doi.org/10.2216/i0031-8884-25-4-551.1
Moissl, C., Osman, S., La Duc, M.T., Dekas, A., Brodie, E., DeSantis, T., Desantis, T. & Venkateswaran, K. (2007) Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS microbiology ecology 61: 509–521. https://doi.org/10.1111/j.1574-6941.2007.00360.x
Mühlsteinová, R. & Hauer, T. (2013) Pilot survey of cyanobacterial diversity from the neighborhood of San Gerardo de Rivas, Costa Rica with a brief summary of current knowledge of terrestrial cyanobacteria in Central America. Brazilian Journal of Botany 36: 299–307. https://doi.org/10.1007/s40415-013-0030-5
Nägeli, C. (1849) Gattungen einzelliger Algen physiologisch und systematisch bearbeitet. Friedrich Schulthess, Zürich. 139 pp. https://doi.org/10.5962/bhl.title.6805
Ortega-Morales, O., Montero-Muñoz, J.L., Baptista Neto, J.A., Beech, I.B., Sunner, J. & Gaylarde, C. (2019) Deterioration and microbial colonization of cultural heritage stone buildings in polluted and unpolluted tropical and subtropical climates: A meta-analysis. International Biodeterioration & Biodegradation 143: 104734. https://doi.org/10.1016/j.ibiod.2019.104734
Osorio-Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Ková?ik, L., Martin, M.P. & Johansen, J.R. (2014) Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. European Journal of Phycology 49: 450–470. https://doi.org/10.1080/09670262.2014.976843
Panwar, P., Allen, M.A., Williams, T.J., Hancock, A.M., Brazendale, S., Bevington, J., Roux, S., Paez-Espino, D., Nayfach, S., Berg, M., Schulz, F., Chen, I.-M.A., Huntemann, M., Shapiro, N., Kyrpides, N.C., Woyke, T., Eloe-Fadrosh, E.A. & Cavicchioli, R. (2020) Influence of the polar light cycle on seasonal dynamics of an Antarctic lake microbial community. Microbiome 8: 116. https://doi.org/10.1186/s40168-020-00889-8
Perkerson III, R.B., Johansen, J.R., Kovácik, L., Brand, J., Kaštovský, J. & Casamatta, D.A. (2011) A unique Pseudanabaenalean (Cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. Journal of Phycology 47: 1397–1412. https://doi.org/10.1111/j.1529-8817.2011.01077.x
Pietrasiak, N., Mühlsteinová, R., Siegesmund, M.A. & Johansen, J.R. (2014) Phylogenetic placement of Symplocastrum (Phormidiaceae, Cyanophyceae) with a new combination S. californicum and two new species: S. flechtnerae and S. torsivum. Phycologia 53: 529–541. https://doi.org/10.2216/14-029.1
Pinheiro, A.C., Mesquita, N., Trovão, J., Soares, F., Tiago, I., Coelho, C., de Carvalho, H.P., Gil, F., Catarino, L., Piñar, G. & Portugal, A. (2019) Limestone biodeterioration: A review on the Portuguese cultural heritage scenario. Journal of Cultural Heritage 36: 275–285. https://doi.org/10.1016/j.culher.2018.07.008
Ribeiro, K.F., Duarte, L. & Crossetti, L.O. (2018) Everything is not everywhere: a tale on the biogeography of cyanobacteria. Hydrobiologia 820: 23–48. https://doi.org/10.1007/s10750-018-3669-x
Rigonato, J., Gama, W.A., Alvarenga, D.O., Branco, L.H.Z., Brandini, F.P., Genuário, D.B. & Fiore, M.F. (2016) Aliterella atlantica gen. nov., sp. nov., and Aliterella antarctica sp. nov., novel members of coccoid Cyanobacteria. International Journal of Systematic and Evolutionary Microbiology 66: 2853–2861. https://doi.org/10.1099/ijsem.0.001066
Rippka, R., Waterbury, J. & Cohen-Bazire, G. (1974) A cyanobacterium which lacks thylakoids. Archives of Microbiology 100: 419–436. https://doi.org/10.1007/BF00446333
Saw, J.H.W., Schatz, M., Brown, M.V., Kunkel, D.D., Foster, J.S., Shick, H., Christensen, S., Hoa, S., Wan, X. & Donachie, S.P. (2013) Cultivation and Complete Genome Sequencing of Gloeobacter kilaueensis sp. nov., from a Lava Cave in Kîlauea Caldera, Hawai’i. PLOS ONE 8: e76376. https://doi.org/10.1371/journal.pone.0076376
Shorthouse, D.P. (2010) SimpleMappr, an online tool to produce publication-quality point maps. Available from: https://www.simplemappr.net/ (Accessed 5 March 2021)
Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. https://doi.org/10.1080/10635150802429642
Tripathi, S.N., Chung, I.K. & Lee, J.A. (2007) Diversity and characteristics of terrestrial cyanobacteria near gimhae city, Korea. Journal of Plant Biology 50: 50–59. https://doi.org/10.1007/BF03030600
Turner, S., Pryer, K.M., Miao, V.P. & Palmer, J.D. (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. The Journal of Eukaryotic Microbiology 46: 327–338. https://doi.org/10.1111/j.1550-7408.1999.tb04612.x
Villa, F. & Cappitelli, F. (2019) The Ecology of Subaerial Biofilms in Dry and Inhospitable Terrestrial Environments. Microorganisms 7: 380. https://doi.org/10.3390/microorganisms7100380
Wang, Y., Cai, F., Jia, N. & Li, R. (2019) Description of a novel coccoid cyanobacterial genus and species Sinocapsa zengkensis gen. nov. sp. nov. (Sinocapsaceae, incertae sedis), with taxonomic notes on genera in Chroococcidiopsidales. Phytotaxa 409: 146–160. https://doi.org/10.11646/phytotaxa.409.3.3
Wilkins, D., Yau, S., Williams, T.J., Allen, M.A., Brown, M.V., DeMaere, M.Z., Lauro, F.M. & Cavicchioli, R. (2013) Key microbial drivers in Antarctic aquatic environments. FEMS Microbiology Reviews 37: 303–335. https://doi.org/10.1111/1574-6976.12007
Zammit, G., Billi, D. & Albertano, P. (2012) The subaerophytic cyanobacterium Oculatella subterranea (Oscillatoriales, Cyanophyceae) gen. et sp. nov.: a cytomorphological and molecular description. European Journal of Phycology 47: 341–354. https://doi.org/10.1080/09670262.2012.717106
Zhang, Q., Zheng, L., Li, T., Li, R. & Song, L. (2018) Aliterella shaanxiensis (Aliterellaceae), a new coccoid cyanobacterial species from China. Phytotaxa 374: 211–220. https://doi.org/10.11646/phytotaxa.374.3.2
Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. https://doi.org/10.1093/nar/gkg595