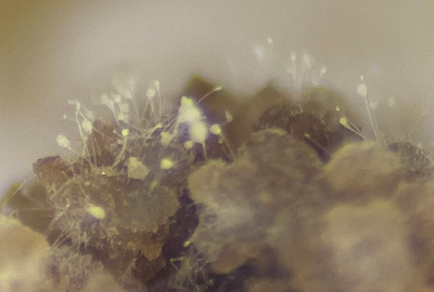
Abstract
Clonostachys xishuangbannaensis is an endophytic fungus isolated from fresh and healthy leaves collected in Xishuangbanna, Yunnan Province, China. It is characterized by solitary, raised, and sparse colonies, mononematous, straight or flexuous, penicillate, and erect conidiophores, and monophialidic, terminal, flask-shaped to cylindrical, aseptate, hyaline phialides. The conidia are single-celled, solitary, acrogenous, simple, and ellipsoidal to oblong with obtuse ends. The species exhibits morphological features typical of the genus Clonostachys. Phylogenetic analyses based on concatenated sequences of the 28S rDNA, translation elongation factor 1-alpha (tef1), RNA polymerase II subunit 2 (rpb2), internal transcribed spacer (ITS), and β-tubulin (tub2) genes, supplemented by morphological assessment, confirmed that C. xishuangbannaensis represents a distinct monophyletic lineage within the genus. Considering the integrated phylogenetic and morphological evidence, we formally propose it as a new species of Clonostachys. This study significantly enhances the understanding of fungal diversity in the previously underexplored region of Yunnan Province, China.
References
- Bao, D.F., Hyde, K.D., Maharachchikumbura, S.S.N., Perera, R.H., Thiyagaraja, V., Hongsanan, S., Wanasinghe, D.N., Shen, H.W., Tian, X.G., Yang, L.Q. & Nalumpang, S. (2023) Taxonomy, phylogeny and evolution of freshwater Hypocreomycetidae (Sordariomycetes). Fungal Diversity 121: 1–94. https://doi.org/10.1007/s13225-023-00521-8
- Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. https://doi.org/10.1080/00275514.1999.12061051
- Clewley, J.P. (1995) Macintosh sequence analysis software - DNAStar’s LaserGene. Molecular Biotechnology 3: 221–224. https://doi.org/10.1007/BF02789332
- Corda, A.C.J. (1839) Icones fungorum hucusque cognitorum, vol. 3.
- Domsch, K.H., Gams, W. & Anderson, T.H. (2007) Compendium of Soil Fungi, 2nd Edn. IHW-Verlag, Eching.
- Dong, W., Hyde, K.D., Jeewon, R., Liao, C.F., Zhao, H.J., Kularathnage, N.D., Li, H., Yang, Y.H., Pem, D., Shu, Y.X., Gafforov, Y., Manawasinghe, I.S. & Doilom, M. (2023) Mycosphere notes 449–468: saprobic and endophytic fungi in China, Thailand, and Uzbekistan. Mycosphere 14 (1): 2208–2262. https://doi.org/10.5943/mycosphere/14/1/26
- Forin, N., Vizzini, A., Nigris, S., Ercole, E., Voyron, S., Girlanda, M. & Baldan, B. (2020) Illuminating type collections of nectriaceous fungi in Saccardo’s fungarium. Persoonia: Molecular Phylogeny and Evolution of Fungi 45: 221–249. https://doi.org/10.3767/persoonia.2020.45.09
- Hall, T. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
- He, S.C., Wei, D.P., Bhunjun, C.S., Zhao, Q. & Jayawardena, R.S. (2023) A new species of Stephanonectria (Bionectriaceae) from southwestern China. Asian Journal of Mycology 6 (1): 98–106. https://doi.org/10.5943/ajom/6/1/9
- He, S.C., Hyde, K.D., Jayawardena, R.S., Thiyagaraja, V., Wanasinghe, D.N., Zhao, Y.W., Wang, Z.Y., Cai, T., Yang, Y.Y., Al-Otibi, F. & Yang, Z. (2025a) Taxonomic contributions to Pleosporales and Kirschsteiniotheliales from the Xizang Autonomous Region, China. Mycology. [38 pp.] https://doi.org/10.1080/21501203.2025.2493072
- He, S.C., Thiyagaraja, V., Bhunjun, C.S., Chomnunti, P., Dissanayake, L.S., Jayawardena, R.S., Yang, H.D., Zhao, Y.W., Al-Otibi, F., Zhao, Q. & Hyde, K.D. (2025b) Morphology and multi-gene phylogeny reveal three new species of Clonostachys and two combinations of Sesquicillium (Bionectriaceae, Hypocreales) from Xizang, China. MycoKeys 115: 43–66. https://doi.org/10.3897/mycokeys.115.139757
- Hirooka, Y. & Kobayashi, T. (2007) Taxonomic studies of nectrioid fungi in Japan. II: The genus Bionectria. Mycoscience 48: 81–89. https://doi.org/10.1007/s10267-006-0331-7
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Hyde, K.D., Norphanphoun, C., Maharachchikumbura, S.S.N., Bhat, D.J., Jones, E.B.G., Bundhun, D., Chen, Y.J., Bao, D.F., Boonmee, S., Calabon, M.S. & Chaiwan, N. (2020a) Refined families of Sordariomycetes. Mycosphere 11: 305–1059. https://doi.org/10.5943/mycosphere/11/1/7
- Hyde, K.D., Jeewon, R., Chen, Y.J., Bhunjun, C.S., Calabon, M.S., Jiang, H.B., Lin, C.G., Norphanphoun, C., Sysouphanthong, P., Jones, E.B.G. & Lumyong, S. (2020b) The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 105: 219–271. https://doi.org/10.1007/s13225-020-00458-2
- Hyde, K.D., Noorabadi, M.T., Thiyagaraja, V., He, M.Q., Johnston, P.R., Wijesinghe, S.N., Armand, A., Biketova, A.Y., Chethana, K.W.T., Erdoğdu, M., Ge, Z.W., Groenewald, J.Z., Hongsanan, S., Kušan, I., Leontyev, D.V., Li, D.W., Lin, C.G., Liu, N.G., Maharachchikumbura, S.S.N., Matočec, N., May, T.W., McKenzie, E.H.C., Mešić, A., Perera, R.H., Phukhamsakda, C., Piątek, M., Samarakoon, M.C., Selcuk, F., Senanayake, I.C., Tanney, J.B., Tian, Q., Vizzini, A., Wanasinghe, D.N., Wannasawang, N., Wijayawardene, N., Zhao, R.L., Abdel-Wahab, M.A., Abdollahzadeh, J., Abeywickrama, P.D., Abhinav, Absalan, S., Acharya, K., Afshari, N., Afshan,. NS, Afzalinia, S., Ahmadpour, S.A., Akulov, O., Alizadeh, A., Alizadeh, M., Al-Sadi, A.M., Alves, A., Alves, V.C.S., Alves-Silva, G., Antonín, V., Aouali, S., Aptroot, A., Apurillo, C.C.S., Arias, R.M., Asgari, B., Asghari, R., Assis, D.M.A., Assyov, B., Atienza, V., Aumentado, H.D.R., Avasthi, S., Azevedo, E., Bakhshi, M., Bao, D.F., Baral, H.O., Barata, M., Barbosa, K.D., Barbosa, R.N., Barbosa, F.R., Baroncelli, R., Barreto, G.G., Baschien, C., Bennett, R.M., Bera, I., Bezerra, J.D.P., Bhunjun, C.S., Bianchinotti, M.V., Błaszkowski, J., Boekhout, T., Bonito, G.M., Boonmee, S., Boonyuen, N., Bortnikov, F.M., Bregant, C., Bundhun, D., Burgaud, G., Buyck, B., Caeiro, M.F., Cabarroi-Hernández, M., Cai, M.F., Cai, L., Calabon, M.S., Calaça, F.J.S., Callalli, M., Câmara, M.P.S., Cano-Lira, J., Cao, B., Carlavilla, J.R., Carvalho, A., Carvalho, T.G., Castañeda-Ruiz, R.F., Catania, M.D.V., Cazabonne, J., Cedeño-Sanchez, M., Chaharmiri-Dokhaharani, S., Chaiwan, N., Chakraborty, N., Cheewankoon, R., Chen, C., Chen, J., Chen, Q., Chen, Y.P., Chinaglia, S., Coelho-Nascimento, C.C., Coleine, C., CostaRezende, D.H., Cortés-Pérez, A., Crouch, J.A., Crous, P.W., Cruz, R.H.S.F., Czachura, P., Damm, U., Darmostuk, V., Daroodi, Z., Das, K., Davoodian, N., Davydov, E.A., da Silva, G.A., da Silva, I.R., da Silva, R.M.F., da Silva, S.A.C., Dai, D.Q., Dai, Y.C., de Groot, M.D., De Kesel, A., De Lange, R., de Medeiros, E.V., de Souza, C.F.A., de Souza, F.A., dela Cruz, T.E.E., Decock, C., Delgado, G., Denchev, C.M., Denchev, T.T., Deng, Y.L., Dentinger, B.T.M., Devadatha, B., Dianese, J.C., Dima, B., Doilom, M., Dissanayake, A.J., Dissanayake, D.M.L.S., Dissanayake, L.S., Diniz, A.G., Dolatabadi, S., Dong, J.H., Dong, W., Dong, Z.Y., Drechsler-Santos, E.R., Druzhinina, I.S., Du, T.Y., Dubey, M.K., Dutta, A.K., Elliott, T.F., Elshahed, M.S., Egidi, E., Eisvand, P., Fan, L., Fan, X., Fan, X.L., Fedosova, A.G., Ferro, L.O., Fiuza, P.O., Flakus, A.W., Fonseca, E.O., Fryar, S.C., Gabaldón, T., Gajanayake, A.J., Gannibal, P.B., Gao, F., GarcíaSánchez, D., García-Sandoval, R., Garrido-Benavent, I., Garzoli, L., Gasca-Pineda, J., Gautam, A.K., Gené, J., Ghobad-Nejhad, M., Ghosh, A., Giachini, A.J., Gibertoni, T.B., Gentekaki, E., Gmoshinskiy, V.I., GóesNeto, A., Gomdola, D., Gorjón, S.P., Goto, B.T., Granados-Montero, M.M., Griffith, G.W., Groenewald, M., Grossart, H.-P., Gu, Z.R., Gueidan, C., Gunarathne, A., Gunaseelan, S., Guo, S.L., Gusmão, L.F.P., Gutierrez, A.C., Guzmán-Dávalos, L., Haelewaters, D., Haituk, H., Halling, R.E., He, S.C., Heredia, G., HernándezRestrepo, M., Hosoya, T., Hoog, S.D., Horak, E., Hou, C.L., Houbraken, J., Htet, Z.H., Huang, S.K., Huang, W.J., Hurdeal, V.G., Hustad, V.P., Inácio, C.A., Janik, P., Jayalal, R.G.U., Jayasiri, S.C., Jayawardena, R.S., Jeewon, R., Jerônimo, G.H., Jin, J., Jones, E.B.G., Joshi, Y., Jurjević, Ž., Justo, A., Kakishima, M., Kaliyaperumal, M., Kang, G.P., Kang, J.C., Karimi, O., Karunarathna, S.C., Karpov, S.A., Kezo, K., Khalid, A.N., Khan, M.K., Khuna, S., Khyaju, S., Kirchmair, M., Klawonn, I., Kraisitudomsook, N., Kukwa, M., Kularathnage, N.D., Kumar, S., Lachance, M.A., Lado, C., Latha, K.P.D., Lee, H.B., Leonardi, M., Lestari, A.S., Li, C., Li, H., Li, J., Li, Q., Li, Y., Li, Y.C., Li, Y.X., Liao, C.F., Lima, J.L.R., Lima, J.M.S., Lima, N.B., Lin, L., Linaldeddu, B.T., Linn, M.M., Liu, F., Liu, J.K., Liu, J.W., Liu, S., Liu, S.L., Liu, X.F., Liu, X.Y., Longcore, J.E., Luangharn, T., Luangsaard, J.J., Lu, L., Lu, Y.Z., Lumbsch, H.T., Luo, L., Luo, M., Luo, Z.L., Ma, J., Madagammana, A.D., Madhushan, A., Madrid, H., Magurno, F., Magyar, D., Mahadevakumar, S., Malosso, E., Malysh, J.M., Mamarabadi, M., Manawasinghe, I.S., Manfrino, R.G., Manimohan, P., Mao, N., Mapook, A., Marchese, P., Marasinghe, D.S., Mardones, M., Marin-Felix, Y., Masigol, H., Mehrabi, M., MehrabiKoushki, M., Meiras-Ottoni, A., de Melo, R.F.R., Mendes-Alvarenga, R.L., Mendieta, S., Meng, Q.F., Menkis, A., Menolli, J.N., Mikšík, M., Miller, S.L., Moncada, B., Moncalvo, J.M., Monteiro, J.S., Monteiro, M., Mora-Montes, H.M., Moroz, E.L., Moura, J.C., Muhammad, U., Mukhopadhyay, S., Nagy, G.L., Najamul, S.A., Najafiniya, M., Nanayakkara, C.M., Naseer, A., Nascimento, E.C.R., Nascimento, S.S., Neuhauser, S., Neves, M.A., Niazi, A.R., Nie, Yong., Nilsson, R.H., Nogueira, P.T.S., Novozhilov, Y.K., Noordeloos, M., Norphanphoun, C., Nuñez, O.N., O’Donnell, R.P., Oehl, F., Oliveira, J.A., Oliveira, J.I., Oliveira, N.V.L., Oliveira, P.H.F., Orihara, T., Oset, M., Pang, K.L., Papp, V., Pathirana, L.S., Peintner, U., Pem, D., Pereira, O.L., Pérez-Moreno, J., Pérez-Ortega, S., Péter, G., Pires-Zottarelli, C.L.A., Phonemany, M., Phongeun, S., Pošta, A., Prazeres, J.F.S.A., Quan, Y., Quandt, C.A., Queiroz, M.B., Radek, R., Rahnama, K., Raj, K.N.A., Rajeshkumar, K.C., Rajwar, S., Ralaiveloarisoa, A.B., Rämä, T., Ramírez-Cruz, V., Rambold, G., Rathnayaka, A.R., Raza, M., Ren, G.C., Rinaldi, A.C., Rivas-Ferreiro, M., Robledo, G.L., Ronikier, A., Rossi, W., Rusevska, K., Ryberg, M., Safi, A., Salimi, F., Salvador-Montoya, C.A., Samant, B., Samaradiwakara, N.P., Sánchez-Castro, I., Sandoval-Denis, M., Santiago, A.L.C.M.A., Santos, A.C.D.S., Santos, L.A., Sarma, V.V., Sarwar, S., Savchenko, A., Savchenko, K., Saxena, R.K., Schoutteten, N., Selbmann, L., Ševčíková, H., Sharma, A., Shen, H.W., Shen, Y.M., Shu, Y.X., Silva, H.F., Silva-Filho, A.G.S., Silva, V.S.H., Simmons, D.R., Singh, R., Sir, E.B., Sohrabi, M., Souza, F.A., Souza-Motta, C.M., Sriindrasutdhi, V., Sruthi, O.P., Stadler, M., Stemler, J., Stephenson, S.L., Stoyneva-Gaertner, M.P., Strassert, J.F.H., Stryjak-Bogacka, M., Su, H., Sun, Y.R., Svantesson, S., Sysouphanthong, P., Takamatsu, S., Tan, T.H., Tanaka, K., Tang, C., Tang, X., Taylor, J.E., Taylor, P.W.J, Tennakoon, D.S., Thakshila, S.A.D., Thambugala, K.M., Thamodini, G.K., Thilanga, D., Thines, M., Tiago, P.V., Tian, X.G., Tian, W.H., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Tomšovský, M., Torruella, G., Tsurykau, A., Udayanga, D., Ulukapı, M., Untereiner, W.A., Usman, M., Uzunov, B.A., Vadthanarat, S., Valenzuela, R., Van den Wyngaert, S., Van Vooren, N., Velez, P., Verma, R.K., Vieira, L.C., Vieira, W.A.S., Vinzelj, J.M., Tang, A.M.C., Walker, A., Walker, A.K., Wang, Q.M., Wang, Y., Wang, X.Y., Wang, Z.Y., Wannathes, N., Wartchow, F., Weerakoon, G., Wei, D.P., Wei, X., White, J.F., Wijesundara, D.S.A., Wisitrassameewong, K., Worobiec, G., Wu, H.X., Wu, N., Xiong, Y.R., Xu, B., Xu, J.P., Xu, R., Xu, R.F., Xu, R.J., Yadav, S., Yakovchenko, L.S., Yang, H.D., Yang, X., Yang, Y.H., Yang, Y., Yang, Y.Y., Yoshioka, R., Youssef, N.H., Yu, F.M., Yu, Z.F., Yuan, L.L., Yuan, Q., Zabin, D.A., Zamora, J.C., Zapata, C.V., Zare, R., Zeng, M., Zeng, X.Y., Zhang, J.F., Zhang, J.Y., Zhang, S., Zhang, X.C., Zhao, C.L., Zhao, H., Zhao, Q., Zhao, H., Zhao, H.J., Zhou, H.M., Zhu, X.Y., Zmitrovich, I.V., Zucconi, L. & Zvyagina, E. (2024) The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 15 (1): 5146–6239. https://doi.org/10.5943/mycosphere/15/1/25
- Jayasiri, S.C., Hyde, K.D., Ariyawansa, H.A., Bhat, J., Buyck, B., Cai, L., Dai, Y.C., Abd-Elsalam, K.A., Ertz, D., Hidayat, I., Jeewon, R., Jones, E.B.G., Bahkali, A.H., Karunarathna, S.C., Liu, J.K., Luangsa-ard, J.J., Lumbsch, H.T., Maharachchikumbura, S.S.N., McKenzie, E.H.C., Moncalvo, J.M., Ghobad-Nejhad, M., Nilsson, H., Pang, K.L., Pereira, O.L., Phillips, A.J.L., Raspé, O., Rollins, A.W., Romero, A.I., Etayo, J., Selçuk, F., Stephenson, S.L., Suetrong, S., Taylor, J.E., Tsui, C.K.M., Vizzini, A., Abdel-Wahab, M.A., Wen, T.C., Boonmee, S., Dai, D.Q., Daranagama, D.A., Dissanayake, A.J., Ekanayaka, A.H., Fryar, S.C., Hongsanan, S., Jayawardena, R.S., Li, W.J., Perera, R.H., Phookamsak, R., de Silva, N.I., Thambugala, K.M., Tian, Q., Wijayawardene, N.N., Zhao, R.L. & Zhao, Q. (2015) The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74 (1): 3–18. https://doi.org/10.1007/s13225-015-0351-8
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2018) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
- Lechat, C., Fournier, J., Chaduli, D., Favel, A. & Lesage-Meessen, L. (2020) Clonostachys saulensis (Bionectriaceae, Hypocreales), a new species from French Guiana. Ascomycete.org 11 (5): 182–186. https://doi.org/10.25664/ART-0260
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Liu, X.F., Tibpromma, S., Hughes, A.C., Chethana, K.W.T., Wijayawardene, N.N., Dai, D.Q., Du, T.Y., Elgorban, A.M., Stephenson, S.L., Suwannarach, N. & Xu, J.C. (2023) Culturable mycota on bats in central and southern Yunnan Province, China. Mycosphere 14: 497–662. https://doi.org/10.5943/mycosphere/14/1/7
- Luo, J. & Zhuang, W.Y. (2007) A new species and two new Chinese records of Bionectria (Bionectriaceae, Hypocreales). Mycotaxon 101: 315–323.
- Luo, J. & Zhuang, W.Y. (2010) Bionectria vesiculosa sp. nov. from Yunnan, China. Mycotaxon 113: 243–249. https://doi.org/10.5248/113.243
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols: a guide to methods and applications. Academic Press, San Diego, pp. 315–322. https://doi.org/10.1109/GCE.2010.5676129
- Moreira, G.M., Abreu, L.M., Carvalho, V.G., Schroers, H.J. & Pfenning, L.H. (2016) Multilocus phylogeny of Clonostachys subgenus Bionectria from Brazil and description of Clonostachys chloroleuca sp. nov. Mycological Progress 15: 1031–1039. https://doi.org/10.1007/s11557-016-1224-6
- Perera, R.H., Hyde, K.D., Maharachchikumbura, S.S.N., Jones, E.B.G., McKenzie, E.H.C., Stadler, M., Lee, H.B., Samarakoon, M.C., Ekanayaka, A.H., Camporesi, E. & Liu, J.K. (2020) Fungi on wild seeds and fruits. Mycosphere 11: 2108–2480. https://doi.org/10.5943/mycosphere/11/1/14
- Perera, R.H., Hyde, K.D., Jones, E.B.G., Maharachchikumbura, S.S.N., Bundhun, D., Camporesi, E., Akulov, A., Liu, J.K. & Liu, Z.Y. (2023) Profile of Bionectriaceae, Calcarisporiaceae, Hypocreaceae, Nectriaceae, Tilachlidiaceae, Ijuhyaceae fam. nov., Stromatonectriaceae fam. nov. and Xanthonectriaceae fam. nov. Fungal Diversity 121: 95–271. https://doi.org/10.1007/s13225-022-00512-1
- Piombo, E., Guaschino, M., Jensen, D.F., Karlsson, M. & Dubey, M. (2023) Insights into the ecological generalist lifestyle of Clonostachys fungi through analysis of their predicted secretomes. Frontiers in Microbiology 14: 1112673. https://doi.org/10.3389/fmicb.2023.1112673
- Prasher, I.B. & Chauhan, R. (2017) Clonostachys indicus sp. nov. from India. Kavaka 48: 22–26.
- Preedanon, S., Suetrong, S., Srihom, C., Somrithipol, S., Kobmoo, N., Saengkaewsuk, S., Srikitikulchai, P., Klaysuban, A., Nuankaew, S., Chuaseeharonnachai, C. & Chainuwong, B. (2023) Eight novel cave fungi in Thailand’s Satun Geopark. Fungal Systematics and Evolution 12: 1–30. https://doi.org/10.3114/fuse.2023.12.01
- Research, M.P. & Service, A.R. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7 (1): 103–116. https://doi.org/10.1006/mpev.1996.0376
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Rossman, A.Y., McKemy, J.M., Pardo-Schultheiss, R.A. & Schroers, H.J. (2001) Molecular studies of the Bionectriaceae using large subunit rDNA sequences. Mycologia 93: 100–110. https://doi.org/10.1080/00275514.2001.12061283
- Schroers, H.J. (2001) A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Studies in Mycology 46: 1–214.
- Tibpromma, S., Hyde, K.D., McKenzie, E.H.C., Bhat, D.J., Phillips, A.J.L., Wanasinghe, D.N., Samarakoon, M.C., Jayawardena, R.S., Dissanayake, A.J., Tennakoon, D.S. & Doilom, M. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. https://doi.org/10.1007/s13225-018-0408-6
- Torcato, C., Gonçalves, M.F.M., Rodríguez-Gálvez, E. & Alves, A. (2020) Clonostachys viticola sp. nov., a novel species isolated from Vitis vinifera. International Journal of Systematic and Evolutionary Microbiology 70 (7): 4321–4328. https://doi.org/10.1099/ijsem.0.004286
- Vaidya, G., Lohman, D.J. & Meier, R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Groenewald, J.Z., Cardinali, G., Houbraken, J., Boekhout, T., Crous, P.W., Robert, V. & Verkley, G.J.M. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. https://doi.org/10.1016/j.simyco.2018.05.001
- Wan, H.C., Yan, F.H., Jian, D.L., Xiao, Z., Zong, Q.L. & Dao, C.J. (2016) A new araneogenous fungus of the genus Clonostachys. Mycosystema 35: 1061–1069. https://doi.org/10.13346/j.mycosystema.150244
- Wang, Y., Tang, D.X., Luo, R., Wang, Y.B., Thanarut, C., Dao, V.M. & Yu, H. (2023) Phylogeny and systematics of the genus Clonostachys. Frontiers in Microbiology 14: 1117753. https://doi.org/10.3389/fmicb.2023.1117753
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols: a guide to methods and applications. Academic Press, San Diego, pp. 315–322.
- Zeng, Z. & Zhuang, W.Y. (2022) Three new species of Clonostachys (Hypocreales, Ascomycota) from China. Journal of Fungi 8 (10): 1–11. https://doi.org/10.3390/jof8101027
- Zhang, Z.Y., Li, X., Chen, W.H., Liang, J.D. & Han, Y.F. (2023) Culturable fungi from urban soils in China II, with the description of 18 novel species in Ascomycota (Dothideomycetes, Eurotiomycetes, Leotiomycetes and Sordariomycetes). MycoKeys 98: 167–220. https://doi.org/10.3897/mycokeys.98.102816
- Zhao, L., Groenewald, J.Z., Hernández-Restrepo, M., Schroers, H.J. & Crous, P.W. (2023) Revising Clonostachys and allied genera in Bionectriaceae. Studies in Mycology 105: 204–265. https://doi.org/10.3114/sim.2023.105.03