Abstract
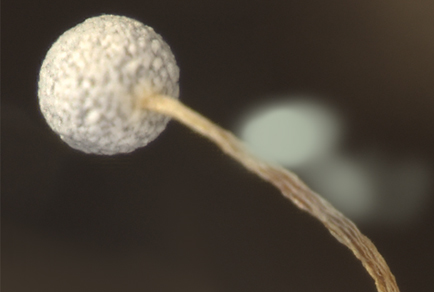
A new species of Physarum discovered in the Trans-Baikal Territory, Russia is described and illustrated. This new species clearly differs from previously known species in several morphological features. The new species and six other species of Physarum (P. bethelii, P. flavicomum, P. galbeum, P. karamanicum, P. oblatum, P. viride) have a very long stalk and threads of capillitium with yellow lime which distinguishes them into a separate morphological group. Sporotheca of the new Physarum are covered with white lime, while the sporotheca of five other species are covered with lime of yellow, orange and green colours. An important distinguishing feature of the new species is the fragile stalk, which breaks off at the top. Sporocarps of the new species were examined by scanning electron microscopy. We present a table of morphological features of Physarum species with long stalks and yellow lime in nodules of threads of capillitium. Based on the sequences of small subunit (18S) of nuclear ribosomal DNA, we reconstructed a phylogeny of the genus Physarum. DNA sequences obtained by us can be used for identification of new species.
References
- Aguilar, M., Fiore-Donno, A.-M., Lado, C. & Cavalier-Smith, T. (2014) Using environmental niche models to test the ‘everything is everywhere’ hypothesis for Badhamia. ISME J 8 (4): 737–745. https://doi.org/10.1038/ismej.2013.183
- Altschul, S., Gish, W., Miller, W., Myers, E. & Lipman, D. (1990) Basic local alignment search tool. Journal of Molecular Biology 215 (3): 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
- Cainelli, R., de Haan, M., Meyer, M., Bonkowski, M. & Fiore-Donno, A.M. (2020) Phylogeny of Physarida (Amoebozoa, Myxogastria) based on the small‐subunit ribosomal RNA gene, redefinition of Physarum pusillum s. str. and reinstatement of P. gravidum Morgan. Journal of Eukaryotic Microbiology 67 (3): 327–336. https://doi.org/10.1111/jeu.12783
- Dagamac, N.H.A., Hoffmann, M., Novozhilov, Yu.K. & Schnittler, M. (2017) Myxomycetes from the highlands of Ethiopia. Nova Hedwigia 104 (1–3): 111–127. https://doi.org/10.1127/nova_hedwigia/2016/0380
- Dai, D., Sun, P., Wu, X., Hu, J., Zhang, B., Wei, Y. & Li, Y. (2023) First report of yellow rot caused by Physarum galbeum in Grifola frondosa in China. Plant Disease 107 (7). https://doi.org/10.1094/PDIS-10-22-2419-PDN
- Felsenstein, J. (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17: 368–376. https://doi.org/10.1007/BF01734359
- Fiore-Donno, A.M., Kamono, A., Meyer, M., Schnittler, M., Fukui, M. & Cavalier-Smith, T. (2012) 18S rDNA phylogeny of Lamproderma and allied genera (Stemonitales, Myxomycetes, Amoebozoa). PLoS One 7 (4): e35359. https://doi.org/10.1371/journal.pone.0035359
- García-Martín, J.M., Mosquera, J. & Lado, C. (2018) Morphological and molecular characterization of a new succulenticolous Physarum (Myxomycetes, Amoebozoa) with unique polygonal spores linked in chains. European Journal of Protistology 63: 13–25. https://doi.org/10.1016/j.ejop.2017.12.004
- García-Martín, J.M., Zamora, J.C. & Lado, C. (2023) Multigene phylogeny of the order Physarales (Myxomycetes, Amoebozoa): shedding light on the dark-spored clade. Persoonia 51 (1): 89–124. https://doi.org/10.3767/persoonia.2023.51.02
- Hoppe, T. & Schnittler, M. (2015) Characterization of myxomycetes in two different soils by TRFLP analysis of partial 18S rRNA gene sequences. Mycosphere 6 (2): 216–227. https://doi.org/10.5943/mycosphere/6/2/11
- Kamono, A., Meyer, M., Cavalier-Smith, T., Fukui, M. & Fiore-Donno, A.M. (2013) Exploring slime mould diversity in high-altitude forests and grasslands by environmental RNA analysis. FEMS Microbiology Ecology 84 (1): 98–109. https://doi.org/10.1111/1574-6941.12042
- Katoh, K. & Standley, D.M. (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30 (4): 772–780. https://doi.org/10.1093/molbev/mst010
- Katoh, K. & Toh, H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9 (4): 286–298. https://doi.org/10.1093/bib/bbn013
- Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. https://doi.org/10.1093/nar/gkf436
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. https://doi.org/10.1093/molbev/msy096
- Lado, C. (2005–2025) An on line nomenclatural information system of Eumycetozoa. Real Jardin Botanico, CSIC. Madrid, Spain. Available from: http://www.nomen.eumycetozoa.com
- Li, X., Hu, J., Tuo, Y., Li, Y., Dai, D., Sossah, F.L., Liu, M., Wang, J., Song, J., Zhang, B., Li, X. & Li, Y. (2024) Catalogue of fungi in China 4: Didymiaceae and Physaraceae (Myxomycetes). Mycology 16 (1): 124–157. https://doi.org/10.1080/21501203.2024.2410508
- Nandipati, S.C.R., Haugli, K., Coucheron, D.H., Haskins, E.F. & Johansen, S.D. (2012) Polyphyletic origin of the genus Physarum (Physarales, Myxomycetes) revealed by nuclear rDNA mini-chromosome analysis and group I intron synapomorphy. BMC Evolutionary Biology 12: 166. https://doi.org/10.1186/1471-2148-12–166
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32 (1): 268–274. https://doi.org/10.1093/molbev/msu300
- Rambaut, A. (2018) FigTree v.1.4.4. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 26 May 2025)
- Shchepin, O.N., Dagamac, N.H., Sanchez, O.M., Novozhilov, Y.K., Schnittler, M. & Zemlyanskaya, I.V. (2017) DNA barcoding as a tool for identification of plasmodia and sclerotia of myxomycetes (Myxogastria) appearing in moist chamber cultures. Mycosphere 8 (10): 1904–1913. https://doi.org/mycosphere/8/10/13
- Shchepin, O.N., Schnittler, M., Erastova, D.A., Prikhodko, I.S., Borg Dahl, M., Azarov, D.V., Chernyaeva, E.N. & Novozhilov, Yu.K. (2019) Community of dark-spored myxomycetes in ground litter and soil of taiga forest (Nizhne-Svirskiy Reserve, Russia) revealed by DNA metabarcoding. Fungal Ecology 39: 80–93. https://doi.org/10.1016/j.funeco.2018.11.006
- Shchepin, O., Novozhilov, Yu., Woyzichovski, J., Bog, M., Prikhodko, I., Fedorova, N., Gmoshinskiy, V., Borg Dahl, M., Dagamac, N.H.A., Yajima, Y. & Schnittler, M. (2022) Genetic structure of the protist Physarum albescens (Amoebozoa) revealed by multiple markers and genotyping by sequencing. Molecular Ecology 31 (1): 372–390. https://doi.org/10.1111/mec.16239
- Stephenson, S.L., Novozhilov, Yu.K. & Prikhodko, I.S. (2020) A new species of Physarum (Myxomycetes) from Christmas Island (Australia). Novosti sistematiki nizshikh rastenii 54 (2): 397–404. https://doi.org/10.31111/nsnr/2020.54.2.397
- Trifinopoulos, J., Nguyen, L.T., von Haeseler, A. & Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44 (W1): 232–235. https://doi.org/10.1093/nar/gkw256