Abstract
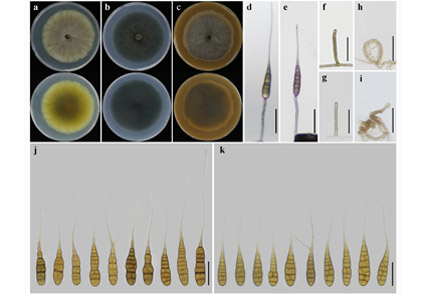
The medicinal plant Atractylodes macrocephala Koidz. (AM) is a common herb in China. In this study, a novel species within the Alternaria genus was isolated from AM plants showing typical Alternaria symptoms in Hefeng County, Enshi Tujia and Miao Autonomous Prefecture of Hubei Province, China. Multi-locus phylogenetic analysis using concatenated sequences of the internal transcribed spacer of the rDNA region (ITS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Alternaria major allergen gene (Alt a 1), translation elongation factor 1 alpha (TEF1), and RNA polymerase second largest subunit (RPB2) genes provided further evidence supporting the placement in Alternaria sect. Porri. Although the newly isolated strains clustered closely with A. guilanica and A. dichondrae, analyses revealed that they belonged to a distinct lineage supporting the establishment of a new species. Based on conidial size, septa, beak morphology, and conidiophore appearance, the isolated strains were differentiated from A. guilanica and A. dichondrae. On the basis of those molecular and morphological characteristics, the fungus is identified as a new species named Alternaria macrocephale.
References
- An, T.J., Park, M.S., Jeong, J.T., Kim, Y.G., Kim, Y.I., Lee, E.S. & Chang, J.K. (2019) Occurrence of the Phytophthora Blight Caused by Phytophthora sansomeana in Atractylodes macrocephala Koidz. Korean Journal of Medicinal Crop Science 27: 404–411.
- https://doi.org/10.7783/KJMCS.2019.27.6.404
- Berbee, M.L., Pirseyedi, M. & Hubbard, S. (1999) Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91: 964–977.
- https://doi.org/10.1080/00275514.1999.12061106
- Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556.
- https://doi.org/10.1080/00275514.1999.12061051
- Editorial Committee of Chinese Pharmacopoeia. (2020) Pharmacopoeia of the People's Republic of China. Beijing: China Medical Science and Technology Press, 107 pp.
- Fan, H., Zang, T., Sheng, J., Zhou, Y. & Kai, G. (2023) First Report of Leaf Spot of Atractylodes macrocephala Caused by Fusarium commune in Zhejiang Province, China. Plant Disease 107: 577.
- https://doi.org/10.1094/PDIS-11-20-2501-PDN
- Gannibal, P.B. (2015) Distribution of Alternaria species among sections. 1. Section Porri. Mycotaxon 130: 207–213.
- https://doi.org/10.5248/130.207
- Gannibal, P.B., Orina, A.S. & Gasich, E.L. (2022) A new section for Alternaria helianthiinficiens found on sunflower and new asteraceous hosts in Russia. Mycological Progress 21: 34.
- https://doi.org/10.1007/s11557-022-01780-6
- Ghafri, A.A., Maharachchikumbura, S., Hyde, K.D., Al-Saady, N.A. & Al-Sadi, A.M. (2019) A new section and a new species of Alternaria encountered from Oman. Phytotaxa 405: 279–289.
- https://doi.org/10.11646/phytotaxa.405.6.1
- Hall, T.A. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
- Hong, S.G., Cramer, R.A., Lawrence, C.B. & Pryor, B.M. (2005) Alt a 1 allergen homologs from Alternaria and related taxa: analysis of phylogenetic content and secondary structure. Fungal Genetics and Biology 42: 119–129.
- https://doi.org/10.1016/j.fgb.2004.10.009
- Huang, X.G., Li, M.Y., Yan, X.N., Yang, J.S., Rao, M.C. & Yuan, X.F. (2021) The potential of Trichoderma brevicompactum for controlling root rot on Atractylodes macrocephala. Canadian Journal of Plant Pathology 43: 794–802.
- https://doi.org/10.1080/07060661.2021.1933602
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Lawrence, D.P., Gannibal, P.B., Dugan, F.M. & Pryor, B.M. (2014) Characterization of Alternaria isolates from the infectoria species-group and a new taxon from Arrhenatherum, Pseudoalternaria arrhenatheria sp. nov. Mycological progress 13: 257–276. https://doi.org/10.1007/s11557-013-0910-x
- Lawrence, D.P., Gannibal, P.B., Peever, T.L. & Pryor, B.M. (2013) The sections of Alternaria: formalizing species-group concepts. Mycologia 105: 530–546. https://doi.org/10.3852/12-249
- Lawrence, D.P., Rotondo, F. & Gannibal, P.B. (2016) Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycological progress 15: 1–22. https://doi.org/10.1007/s11557-015-1144-x
- Li, J.F., Jiang, H.B., Jeewon, R, Hongsanan, S, Bhat, D.J., Tang, S.M., Lumyong, S, Mortimer, P.E., Xu, J.C., Camporesi, E, Bulgakov, T.S., Zhao, G.J., Suwannarach, N. & Phookamsak, R. (2023) Alternaria: Update on species limits, evolution, multi-locus phylogeny, and classification. Studies in Fungi 8: 1–61 https://doi.org/10.48130/SIF-2023-0001
- Liu, H.F., Liao, J., Chen, X.Y., Liu, Q.K., Yu, Z.H. & Deng, J.X. (2019) A novel species and a new record of Alternaria isolated from two Solanaceae plants in China. Mycological Progress 18: 1005–1012. https://doi.org/10.1007/s11557-019-01504-3
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Nees von Esenbeck, C.G. (1816) Das System der Pilze und Schwamme. Stahelschen Buchhandlung, Wurzburg, Germany, 329 pp.
- https://doi.org/10.5962/bhl.title.110007
- Nwe, Z.M., Htut, K.N., Aung, S.L.L., Gou, Y.N., Huang, C.X. & Deng, J.X. (2024) Two novel species and a new host record of Alternaria (Pleosporales, Pleosporaceae) from sunflower (Compositae) in Myanmar. MycoKeys 105: 337–354. https://doi.org/10.3897/mycokeys.105.123790
- Nylander, J.A.A. (2004) MrModeltest v2.0. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- Pei, D.F., Aung, S.L.L., Liu, H.F., Liu, Q.K., Yu, Z.H. & Deng, J.X. (2019) Alternaria hydrangeae sp. nov. (Ascomycota: Pleosporaceae) from Hydrangea paniculata in China. Phytotaxa 401: 287–295. https://doi.org/10.11646/phytotaxa.401.4.6
- Posada, D. & Crandall, K.A. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics (Oxford, England) 14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817
- Poursafar, A., Hashemlou, E., Ghosta, Y., Salimi, F. & Javan-Nikkhah, M. (2021) Alternaria guilanica sp. nov., a new fungal pathogen causing leaf spot and blight on eggplant in Iran. Phytotaxa 520: 184−94. https://doi.org/10.11646/phytotaxa.520.2.5
- Pryor, B.M. & Bigelow, D.M. (2003) Molecular characterization of Embellisia and Nimbya species and their relationship to Alternaria, Ulocladium and Stemphylium. Mycologia 95: 1141–1154. https://doi.org/10.1080/15572536.2004.11833024
- Pryor, B.M. & Gilbertson, R.L. (2000) Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mt SSU rDNA sequences. Mycological Research 104: 1312–1321. https://doi.org/10.1017/S0953756200003002
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Runa, F., Park, M.S. & Pryor, B.M. (2009) Ulocladium systematics revisited: phylogeny and taxonomic status. Mycological progress 8: 35–47. https://doi.org/10.1007/s11557-008-0576-y
- Simmons, E.G. (2000) Alternaria themes and variations. Species on Solanaceae. Mycotaxon 75: 1–115.
- Simmons, E.G. (2007) Alternaria: An identification manual, CBS Biodiversity Series 6. Utrecht: Centraalbureau voor Schimmelcultures.
- Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758−71. https://doi.org/10.1080/10635150802429642
- Swofford, D.L. (2002) PAUP, Phylogenetic analysis using parsimony (and other methods). Version 4.0b10. Sinauer Associates, Sunderland, MA.
- Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38: 3022–3027. https://doi.org/10.1093/molbev/msab120
- Tan, G.Y., Yang, Z.L., Yuan, Z.L. & Zhang, S.A. (2013) Morphological, molecular and pathogenic characterization of Alternaria longipes, the fungal pathogen causing leaf spot on Atractylodes macrocephala. African Journal of Microbiology Research 7: 2589–2595. https://doi.org/10.5897/AJMR12.2091
- Watanabe, M., Lee, K., Goto, K., Kumagai, S., Sugita-Konishi, Y. & Hara-Kudo, Y. (2010) Rapid and effective DNA extraction method with bead Grinding for a large amount of fungal DNA. Journal of Food Protection 73: 1077–1084. https://doi.org/10.4315/0362-028X-73.6.1077
- Wang, Y.X., Jia, F.J., Li, Y. & Huang, J.B. (2007) Biological characteristics of the pathogen causing dark leaf spot on Atractylodes lancea. Plant Protection 33: 107–110.
- White, T.J., Bruns, T., Lee, S.J.W.T. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, San Diego, ifornia, USA, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Woudenberg, J.H.C., Groenewald, J.Z., Binder, M. & Crous, P.W. (2013) Alternaria redefined. Studies in mycology 75: 171–212. https://doi.org/10.3114/sim0015
- Woudenberg, J.H.C., Seidl, M.F., Groenewald, J.Z., De Vries, M., Stielow, J.B., Thomma, B.P.H.J. & Crous, P.W. (2015) Alternaria section Alternaria: Species, formae speciales or pathotypes? Studies in mycology 82: 1–21. https://doi.org/10.1016/j.simyco.2015.07.001
- Woudenberg, J.H.C., Truter, M., Groenewald, J.Z. & Crous, P.W. (2014) Large-spored Alternaria pathogens in section Porri disentangled. Studies in Mycology 79: 1–47. https://doi.org/10.1016/j.simyco.2014.07.003
- Yang, R., Fan, H., He, B., Ruan, Q., Wei, B., Han, B., Hao, X., Maoz, I. & Kai, G. (2023) Current progress of Atractylodes macrocephala Koidz.: A review of its biogeography, PAO-ZHI processing, biological activities, biosynthesis pathways, and technology applications. Medicinal Plant Biology 2. https://doi.org/10.48130/MPB-2023-0005
- You, J.M., Lin, X.M., Guo, J., Zhang, M.D., Liao, C.L., He, M.J., You, J.W. & Sun, Y.L. (2013) First report of root rot on Atractylodes macrocephala (Largehead Atractylodes Rhizome) caused by Ceratobasidium sp. in China. Plant Disease 97: 139–139. https://doi.org/10.1094/PDIS-05-12-0467-PDN
- Zhang, T.Y. (2003) Flora Fungorum Sinicorum-Alternaria 16: 32−86.
- Zhao, L., Luo, H., Cheng, H., Gou, Y.N., Yu, Z.H. & Deng, J.X. (2022) New species of large-spored Alternaria in section porri associated with Compositae plants in China. Journal of Fungi 8: 607. https://doi.org/10.3390/jof8060607
- Zhu, B., Zhang, Q.L., Hua, J.W., Cheng, W.L. & Qin, L.P. (2018) The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review. Journal of Ethnopharmacology 226: 143−67. https://doi.org/10.1016/j.jep.2018.08.023