Abstract
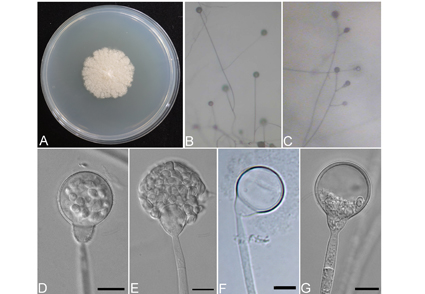
Rhizosphere fungi play crucial roles in plant health and soil ecology. We describe and illustrate Gongronella fusoacuminata, a new species isolated from the rhizosphere soil of pine and mallow in Taiwan. Phylogenetic analyses based on both ITS and LSU rDNA sequences, in conjunction with unique morphological characteristics, confirmed its distinct taxonomic classification within the genus. The phylogenetic analyses revealed that G. fusoacuminata possesses distinctive sequences, clearly differentiating it from other species within the genus. In addition, G. fusoacuminata is distinguished by its fusiform sporangiospores with acuminate apices and their relatively large size, clearly discerning it from other documented species.
References
- Adamčík, S., Cai, L., Chakraborty, D., Chen, X.-H., Cotter, H.V.T., Dai, D.Q., Dai, Y.-C., Das, K., Deng, C., Ghobad-Nejhad, M., Hyde, K.D., Langer, E., Latha, K.P.D., Liu, F., Liu, S.-L., Liu, T., Lv, W., Lv, S.-X., Machado, A.R., Pinho, D.B., Pereira, O.L., Prasher, I.B., Rosado, A.W.C., Qin, J., Qin, W.-M., Verma, R.K., Wang, Q., Yang, Z.-L., Yu, X.-D., Zhou, L.-W. & Buyck, B. (2015) Fungal biodiversity profiles 1–10. Cryptogamie, Mycologie 36: 121–166. https://doi.org/10.7872/crym/v36.iss2.2015.121
- Ariyawansa, H.A., Hyde, K.D., Jayasiri, S.C., Buyck, B., Chethana, K.W.T., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Jayawardena, R.S., Lücking, R., Ghobad-Nejhad, M., Niskanen, T., Thambugala, K.M., Voigt, K., Zhao, R.L., Li, G.-J., Doilom, M., Boonmee, S., Yang, Z.L., Cai, Q., Cui, Y.-Y., Bahkali, A.H., Chen, J., Cui, B.K., Chen, J.J., Dayarathne, M.C., Dissanayake, A.J., Ekanayaka, A.H., Hashimoto, A., Hongsanan, S., Jones, E.B.G., Larsson, E., Li, W.J., Li, Q.-R., Liu, J.K., Luo, Z.L., Maharachchikumbura, S.S.N., Mapook, A., McKenzie, E.H.C., Norphanphoun, C., Konta, S., Pang, K.L., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Pinruan, U., Randrianjohany, E., Singtripop, C., Tanaka, K., Tian, C.M., Tibpromma, S., Abdel-Wahab, M.A., Wanasinghe, D.N., Wijayawardene, N.N., Zhang, J.-F., Zhang, H., Abdel-Aziz, F.A., Wedin, M., Westberg, M., Ammirati, J.F., Bulgakov, T.S., Lima, D.X., Callaghan, T.M., Callac, P., Chang, C.-H., Coca, L.F., Dal-Forno, M., Dollhofer, V., Fliegerová, K., Greiner, K., Griffith, G.W., Ho, H.-M., Hofstetter, V., Jeewon, R., Kang, J.C., Wen, T.-C., Kirk, P.M., Kytövuori, I., Lawrey, J.D., Xing, J., Li, H., Liu, Z.Y., Liu, X.Z., Liimatainen, K., Lumbsch, H.T., Matsumura, M., Moncada, B., Nuankaew, S., Parnmen, S., de Azevedo Santiago, A.L.C.M., Sommai, S., Song, Y., de Souza, C.A.F., de Souza-Motta, C.M., Su, H.Y., Suetrong, S., Wang, Y., Wei, S.-F., Wen, T.C., Yuan, H.S., Zhou, L.W., Réblová, M., Fournier, J., Camporesi, E., Luangsa-ard, J.J., Tasanathai, K., Khonsanit, A., Thanakitpipattana, D., Somrithipol, S., Diederich, P., Millanes, A.M., Common, R.S., Stadler, M., Yan, J.Y., Li, X.H., Lee, H.W., Nguyen, T.T.T., Lee, H.B., Battistin, E., Marsico, O., Vizzini, A., Vila, J., Ercole, E., Eberhardt, U., Simonini, G., Wen, H.-A., Chen, X.-H., Miettinen, O., Spirin, V. & Hernawati (2015) Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75: 27–274. https://doi.org/10.1007/s13225-015-0346-5
- Breidenbach, B., Pump, J. & Dumont, M.G. (2016) Microbial community structure in the rhizosphere of rice plants. Frontiers in Microbiology 6: 1537. https://doi.org/10.3389/fmicb.2015.01537
- Chernomor, O., von Haeseler, A. & Minh, B.Q. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. https://doi.org/10.1093/sysbio/syw037
- Dannaoui, E. (2009) Molecular tools for identification of Zygomycetes and the diagnosis of zygomycosis. Clinical Microbiology and Infection 15: 66–70. https://doi.org/10.1111/j.1469-0691.2009.02983.x
- Dong, C.-B., Zhang, Z.-Y., Chen, W.-H., Han, Y.-F., Huang, J.-Z. & Liang, Z.-Q. (2019) Gongronella zunyiensis sp. nov. (Cunninghamellaceae, Mucorales) isolated from rhizosphere soil in China. Phytotaxa 425 (5): 290–296. https://doi.org/10.11646/phytotaxa.425.5.4
- Hesseltine, C.W. & Ellis, J.J. (1961) Notes on Mucorales, especially Absidia. Mycologia 53: 406–426. https://doi.org/10.2307/3756584
- Hoang, D.T., Chernomor, O., von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281
- de Hoog, G.S. & Gerrits van den Ende, A.H. (1998) Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41: 183–189. https://doi.org/10.1111/j.1439-0507.1998.tb00321.x
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
- Li, G.J., Hyde, K.D., Zhao, R.L., Hongsanan, S., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Alvarado, P., Alves-Silva, G., Ammirati, J.F., Ariyawansa, H.A., Baghela, A., Bahkali, A.H., Beug, M., Bhat, D.J., Bojantchev, D., Boonpratuang, T., Bulgakov, T.S., Camporesi, E., Boro, M.C., Ceska, O., Chakraborty, D., Chen, J.J., Chethana, K.W.T., Chomnunti, P., Consiglio, G., Cui, B.K., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Das, K., Dayarathne, M.C., De Crop, E., De Oliveira, R.J.V., de Souza, C.A.F., de Souza, J.I., Dentinger, B.T.M., Dissanayake, A.J., Doilom, M., Drechsler-Santos, E.R., Ghobad-Nejhad, M., Gilmore, S.P., Góes-Neto, A., Gorczak, M., Haitjema, C.H., Hapuarachchi, K.K., Hashimoto, A., He, M.Q., Henske, J.K., Hirayama, K., Iribarren, M.J., Jayasiri, S.C., Jayawardena, R.S., Jeon, S.J., Jerônimo, G.H., Jesus, A.L., Jones, E.B.G., Kang, J.C., Karunarathna, S.C., Kirk, P.M., Konta, S., Kuhnert, E., Langer, E., Lee, H.S., Lee, H.B., Li, W.J., Li, X.H., Liimatainen, K., Lima, D.X., Lin, C.G., Liu, J.K., Liu, X.Z., Liu, Z.Y., Luangsa-ard, J.J., Lücking, R., Lumbsch, H.T., Lumyong, S., Leaño, E.M., Marano, A.V., Matsumura, M., McKenzie, E.H.C., Mongkolsamrit, S., Mortimer, P.E., Nguyen, T.T.T., Niskanen, T., Norphanphoun, C., O’Malley, M.A., Parnmen, S., Pawłowska, J., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Pires-Zottarelli, C.L.A., Raspé, O., Reck, M.A., Rocha, S.C.O., de Santiago, A.L.C.M.A., Senanayake, I.C., Setti, L., Shang, Q.J., Singh, S.K., Sir, E.B., Solomon, K.V., Song, J., Srikitikulchai, P., Stadler, M., Suetrong, S., Takahashi, H., Takahashi, T., Tanaka, K., Tang, L.P., Thambugala, K.M., Thanakitpipattana, D., Theodorou, M.K., Thongbai, B., Thummarukcharoen, T., Tian, Q., Tibpromma, S., Verbeken, A., Vizzini, A., Vlasák, J., Voigt, K., Wanasinghe, D.N., Wang, Y., Weerakoon, G., Wen, H.A., Wen, T.C., Wijayawardene, N.N., Wongkanoun, S., Wrzosek, M., Xiao, Y.P., Xu, J.C., Yan, J.Y., Yang, J., Yang, S.D., Yu, H., Zhang, J.F., Zhao, J., Zhou, L.W., Peršoh, D., Phillips, A.J.L. & Maharachchikumbura, S.S.N. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. https://doi.org/10.1007/s13225-016-0366-9
- Martins, M.R., Santos, C., Soares, C., Santos, C. & Lima, N. (2020) Gongronella eborensis sp. nov. from vineyard soil of Alentejo (Portugal). International Journal of Systematic and Evolutionary Microbiology 70: 3475–3482. https://doi.org/10.1099/ijsem.0.004201
- Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. https://doi.org/10.1093/molbev/msaa015
- Obase, K., Kitagami, Y., Tanikawa, T., Chen, C.-F. & Matsuda, Y. (2023) Fungi and bacteria in the rhizosphere of Cryptomeria japonica exhibited different community assembly patterns at regional scales in East Asia. Rhizosphere 28: 100807. https://doi.org/10.1016/j.rhisph.2023.100807
- O'Donnell, K. (1993) Fusarium and its near relatives. In: Reynolds, D.R. & Taylor, J.W. (Eds.) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp. 225–233. [https://www.researchgate.net/publication/224900762_Fusarium_and_its_near_relatives]
- Peyronel, B. & Dal Vesco, G. (1955) Ricerche sulla microflora di un terreno agrario presso Torino. Allionia 2: 357–417.
- Ribaldi, M.S. (1952) Sopra un interessante Zigomicete terricola: Gongronella urceolifera n. gen. et n. sp. Rivista di Biologia 44: 157–166.
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Schoch, C.L., Seifert, K.A., Huhndorf, S., Robert, V., Spouge, J.L., Levesque, C.A. & Chen, W. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proceedings of the National Academy of Sciences of the United States of America 109: 6241–6246. https://doi.org/10.1073/pnas.1117018109
- Schwarz, P., Bretagne, S., Gantier, J.C., Garcia-Hermoso, D., Lortholary, O., Dromer, F. & Dannaoui, E. (2006) Molecular identification of zygomycetes from culture and experimentally infected tissues. Journal of Clinical Microbiology 44: 340–349. https://doi.org/10.1128/JCM.44.2.340-349.2006
- Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38: 3022–3027. https://doi.org/10.1093/molbev/msab120
- Tibpromma, S., Hyde, K.D., Jeewon, R., Maharachchikumbura, S.S.N., Liu, J.-K., Bhat, D.J., Jones, E.B.G., McKenzie, E.H.C., Camporesi, E., Bulgakov, T.S., Doilom, M., de Azevedo Santiago, A.L.C.M., Das, K., Manimohan, P., Gibertoni, T.B., Lim, Y.W., Ekanayaka, A.H., Thongbai, B., Lee, H.B., Yang, J.-B., Kirk, P.M., Sysouphanthong, P., Singh, S.K., Boonmee, S., Dong, W., Raj, K.N.A., Latha, K.P.D., Phookamsak, R., Phukhamsakda, C., Konta, S., Jayasiri, S.C., Norphanphoun, C., Tennakoon, D.S., Li, J., Dayarathne, M.C., Perera, R.H., Xiao, Y., Wanasinghe, D.N., Senanayake, I.C., Goonasekara, I.D., de Silva, N.I., Mapook, A., Jayawardena, R.S., Dissanayake, A.J., Manawasinghe, I.S., Chethana, K.W.T., Luo, Z.-L., Hapuarachchi, K.K., Baghela, A., Soares, A.M., Vizzini, A., Meiras-Ottoni, A., Mešić, A., Dutta, A.K., de Souza, C.A.F., Richter, C., Lin, C.-G., Chakrabarty, D., Daranagama, D.A., Lima, D.X., Chakraborty, D., Ercole, E., Wu, F., Simonini, G., Vasquez, G., da Silva, G.A., Plautz Jr., H.L., Ariyawansa, H.A., Lee, H., Kušan, I., Song, J., Sun, J., Karmakar, J., Hu, K., Semwal, K.C., Thambugala, K.M., Voigt, K., Acharya, K., Rajeshkumar, K.C., Ryvarden, L., Jadan, M., Hosen, M.I., Mikšík, M., Samarakoon, M.C., Wijayawardene, N.N., Kim, N.K., Matočec, N., Singh, P.N., Tian, Q., Bhatt, R.P., de Oliveira, R.J.V., Tulloss, R.E., Aamir, S., Kaewchai, S., Marathe, S.D., Khan, S., Hongsanan, S., Adhikari, S., Mehmood, T., Bandyopadhyay, T.K., Svetasheva, T.Y., Nguyen, T.T.T., Antonín, V., Li, W.-J., Wang, Y., Indoliya, Y., Tkalčec, Z., Elgorban, A.M., Bahkali, A.H., Tang, A.M.C., Su, H.-Y., Zhang, H., Promputtha, I., Luangsa-ard, J., Xu, J., Yan, J., Ji-Chuan, K., Stadler, M., Mortimer, P.E., Chomnunti, P., Zhao, Q., Phillips, A.J.L., Nontachaiyapoom, S., Wen, T.-C. & Karunarathna, S.C. (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83: 1–261. https://doi.org/10.1007/s13225-017-0378-0
- Upadhyay, H.P. (1969) Soil fungi from north-east and north Brazil—VII. The genus Gongronella. Nova Hedwigia 17: 65–73. [https://batista.fungibrasil.net/pdf/IMUR631.pdf]
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246
- Vitale, R.G., de Hoog, G.S., Schwarz, P., Dannaoui, E., Deng, S., Machouart, M., Voigt, K., van de Sande, W.W., Dolatabadi, S., Meis, J.F. & Walther, G. (2012) Antifungal susceptibility and phylogeny of opportunistic members of the order Mucorales. Journal of Clinical Microbiology 50: 66–75. https://doi.org/10.1128/JCM.06133-11
- Walther, G., Pawlowska, J., Alastruey-Izquierdo, A., Wrzosek, M., Rodriguez-Tudela, J.L., Dolatabadi, S., Chakrabarti, A. & de Hoog, G.S. (2013) DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30: 11–47. https://doi.org/10.3767/003158513X665070
- Walther, G., Wagner, L. & Kurzai, O. (2019) Updates on the taxonomy of Mucorales with an emphasis on clinically important taxa. Journal of Fungi 5: 106. https://doi.org/10.3390/jof5040106
- Wang, Y.X., Zhao, H., Ding, Z.Y., Ji, X.Y., Zhang, Z.X., Wang, S., Zhang, X.G. & Liu, X.Y. (2023) Three new species of Gongronella (Cunninghamellaceae, Mucorales) from soil in Hainan, China based on morphology and molecular phylogeny. Journal of Fungi (Basel) 9: 1182. https://doi.org/10.3390/jof9121182
- White, T.J., Bruns, T.D., Lee, S.B. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols—a guide to methods and applications. Academic Press, San Diego, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Zhang, Z.-Y., Han, Y.-F., Chen, W.-H. & Liang, Z.-Q. (2019) Gongronella sichuanensis (Cunninghamellaceae, Mucorales), a new species isolated from soil in China. Phytotaxa 416: 167–174. https://doi.org/10.11646/phytotaxa.416.2.4