Abstract
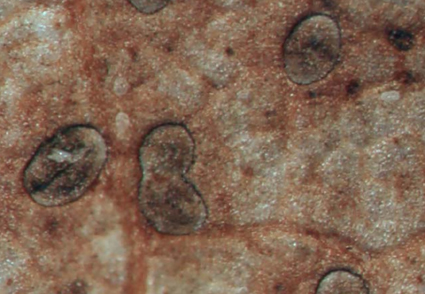
Specimens of Terriera collected from dead leaves of woody plant in the Fodingshan National Nature Reserve, Guizhou Province, China, were identified as a new species and named Terriera tongrenensis. Multi-gene phylogenetic analyses based on ITS-nrLSU-mtSSU show that T. tongrenensis clusters with other Terriera species and is closely related to Terriera houjiazhuangensis, Terriera guizhouensis, Terriera pandanicola, and Terriera sigmoideospora. Morphological comparison reveals distinct differences between the new species and its relatives. T. tongrenensis has the ascomata on abaxial surface of dead leaves, the covering stroma consisting of an innermost layer with a few thin-walled, hyaline, angular to globose cells near the opening and extending to the basal stroma, the absence of an excipulum and the internal matrix of stroma. These findings highlight the unique characteristics of T. tongrenensis, reinforcing its status as a distinct species within the genus.
References
- Cai, S.R., Wang, S.J., Lv, T. & Hou, C.L. (2020) Three new species of Terriera (Rhytismatales, Ascomycota) from China. Mycological Progress 19: 825–835. https://doi.org/10.1007/s11557-020-01594-4
- Eriksson, B. (1970) On Ascomycetes on Diapensales and Ericales in Fennoscandia. 1. Discomycetes. Symbolae Botanicae Upsalienses 19: 1–71.
- Gardes, M. & Bruns, T.D. (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
- Guo, M.J., Zhuo, L., Wang, S.J., Sui, X.N., Zhou, H., Cai, S.R., Luo, J.T., Lei, R.H., Shen, X.Y., Piepenbring, M. & Hou, C.L. (2024) Hyperdiverse Rhytismatales on twigs of Rhododendron spp. Mycosphere 15 (1): 764–880. https://doi.org/10.5943/mycosphere/15/1/6
- Haelewaters, D., Dima, B., Abdel-Hafiz, A.I., Abdel-Wahab, M.H., Abul-ezz, S.R., Acar, İ., Aguirre-Acosta, E., Aime, C., Aldemir, S., Ali, M., Ayala-Vásquez, O., Bakhit, M.S., Bashir, H., Battistin, E., Bendiksen, E., Castro-Rivera, R., Çolak, Ö.F., Kesel, A.D., Fuente, J.I., Di̇zkirici, A., Hussain, S., Jansen, G.M., Kaygusuz, O., Khalid, A.N., Khan, J., Kiyashko, A.A., Larsson, E., Martínez-González, C.R., Morozova, O.V., Niazi, A.R., Noordeloos, M.E., Pham, T.H., Popov, E., Psurtseva, N.V., Schoutteten, N., Sher, H., Türkekul, İ., Verbeken, A., Ahmad, H., Afshan, N.U., Christe, P., Fiaz, M., Glaizot, O., Liu, J.N., Majeed, J., Markotter, W., Nagy, A.A., Nawaz, H., Papp, V., Péter, Á., Pfliegler, W.P., Qasim, T., Riaz, M., Sándor, A.D., Szentiványi, T., Voglmayr, H., Yousaf, N. & Krisai-Greilhuber, I. (2020) Fungal Systematics and Evolution: FUSE 6. Sydowia 72. https://doi.org/10.12905/0380.SYDOWIA72-2020-0231
- Han, J.G., Hosoya, T., Sung, G.H. & Shin, H.D. (2014) Phylogenetic reassessment of Hyaloscyphaceae sensu lato (Helotiales, Leotiomycetes) based on multigene analyses. Fungal Biology 118: 150–167. https://doi.org/10.1016/j.funbio.2013.11.004
- Hou, C.L., Li, L. & Piepenbring, M. (2009) Lophodermium pini-mugonis sp. nov. on needles of Pinus mugo from the Alps based on morphological and molecular data. Mycological Progress 8: 29–33. https://doi.org/10.1007/s11557-008-0575-z
- Hou, C.L., Trampe, T. & Piepenbring, M. (2010) A new species of Rhytisma causes Tar Spot on Comarostaphylis arbutoides (Ericaceae) in Panama. Mycopathologia 169: 225–229. https://doi.org/10.1007/s11046-009-9250-4
- Huelsenbeck, J.P., Ronquist, F., Nielsen, R. & Bollback, J.P. (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310–2314. https://doi.org/10.1126/science.1065889
- Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., McKenzie, E.H., Jones, E.B., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Banmai, S., Chomnunti, P., Cui, B., Daranagama, D.A., Das, K., Dayarathne, M.C., DE SILVA, N.I., Dissanayake, A.J., Doilom, M., Ekanayaka, A.H., Gibertoni, T.B., Góes-Neto, A., Huang, S., Jayasiri, S.C., Jayawardena, R.S., Konta, S., Lee, H.B., Li, W., Lin, C., Liu, J., Lu, Y., Luo, Z., Manawasinghe, I.S., Manimohan, P., Mapook, A., Niskanen, T., Norphanphoun, C., Papizadeh, M., Perera, R.H., Phukhamsakda, C., Richter, C., de A. Santiago, A., Drechsler-Santos, E.R., Senanayake, I.C., Tanaka, K., Tennakoon, T.M., Thambugala, K.M., Tian, Q., Tibpromma, S., Thongbai, B., Vizzini, A., Wanasinghe, D.N., Wijayawardene, N.N., Wu, H., Yang, J., Zeng, X., Zhang, H., Zhang, J., Bulgakov, T.S., Camporesi, E., Bahkali, A.H., Amoozegar, M.A., Araujo-Neta, L.S., Ammirati, J.F., Baghela, A., Bhatt, R.P., Bojantchev, D., Buyck, B., da Silva, G.A., de Lima, C.L., de Oliveira, R., de Souza, C.A., Dai, Y., Dima, B., Duong, T.T., Ercole, E., Mafalda-Freire, F., Ghosh, A., Hashimoto, A., Kamolhan, S., Kang, J., Karunarathna, S.C., Kirk, P.M., Kytövuori, I., Lantieri, A., Liimatainen, K., Liu, Z., Liu, X.Z., Lücking, R., Medardi, G., Mortimer, P.E., Nguyen, T.T., Promputtha, I., Raj, K.N., Reck, M.A., Lumyong, S., Shahzadeh-Fazeli, S.A., Stadler, M., Soudi, M.R., Su, H., Takahashi, T., Tangthirasunun, N., Uniyal, P., Wang, Y., Wen, T., Xu, J.C., Zhang, Z., Zhao, Y., Zhou, J. & Zhu, L. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x
- Johnston, P.R. (1988) An undescribed pattern of ascocarp development in some non-coniferous Lophodermium species. Mycotaxon 31: 383–394.
- Johnston, P.R. (1989) Lophodermium (Rhytismataceae) on Clusia. Sydowia 41: 170–179.
- Johnston, P.R., Quijada, L., Smith, C.A., Baral, H.O., Hosoya, T., Baschien, C., Pärtel, K., Zhuang, W.Y., Haelewaters, D., Park, D., Carl, S., López-Giráldez, F., Wang, Z. & Townsend, J.P. (2019) A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA fungus 1: 1–22. https://doi.org/10.1186/s43008-019-0002-x
- Karakehian, J.M., Quijada, L., Friebes, G., Tanney, J.B. & Pfister, D.H. (2019) Placement of Triblidiaceae in Rhytismatales and comments on unique ascospore morphologies in Leotiomycetes (Fungi, Ascomycota). MycoKeys 54: 99−133. https://doi.org/10.3897/mycokeys.54.35697
- Kirk, P.M., Cannon, P.F., Minter, D.W. & Stalpers, J.A. (2008) Ainsworth & Bisby’s Dictionary of the Fungi. 10th edn. Wallingford, 771 pp. https://doi.org/10.1016/j.mycres.2009.07.003
- Lantz, H., Johnston, P.R., Park, D. & Minter, D.W. (2011) Molecular phylogeny reveals a core clade of Rhytismatales. Mycologia 103: 57–74. https://doi.org/10.3852/10-060
- Li, Q., Wu, Y., Lu, D.D., Xu, Y.F. & Lin, Y.R. (2015) A new species of Terriera (Rhytismatales, Ascomycota) on Photinia villosa. Mycotaxon 130: 27–31. https://doi.org/10.5248/130.27
- Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. https://doi.org/10.1093/molbev/msaa015
- Rambaut, A. (2000) Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16: 395–399. https://doi.org/10.1093/bioinformatics/16.4.395
- Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian Phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
- Spatafora, J.W., Sung, G.H., Johnson, D., Hesse, C., O’Rourke, B., Serdani, M., Spotts, R., Lutzoni, F., Hofstetter, V., Miadlikowska, J., Reeb, V., Gueidan, C., Fraker, E., Lumbsch, T., Lücking, R., Schmitt, I., Hosaka, K., Aptroot, A., Roux, C., Miller, A.N., Geiser, D.M., Hafellner, J., Hestmark, G., Arnold, A.E., Büdel, B., Rauhut, A., Hewitt, D., Untereiner, W.A., Cole, M.S., Scheidegger, C., Schultz, M., Sipman, H. & Schoch, C.L. (2006) A five-gene phylogeny of Pezizomycotina. Mycologia 98 (6): 1018–1028. https://doi.org/10.3852/mycologia.98.6.1018
- Sui, X.N., Guo, M.J., Zhou, H. & Hou, C.L. (2023) Four new species of Phyllosticta from China based on morphological and phylogenetic characterization. Mycology 14 (3): 190–203. https://doi.org/10.1080/21501203.2023.2225552
- Swofford, D.L. (1998) PAUP*: phylogenetic analysis using parsimony (and other methods). Version 4. Sunderland, Massachusetts: Sinauer Associates.
- Tian, H.Z., Yang, Z., Wang, S., Hou, C.L. & Piepenbring, M. (2013) A new species and phylogenetic data for Nematococcomyces. Botany 91 (9): 592–596. https://doi.org/10.1139/cjb-2012-0306
- Tibpromma, S., Hyde, K.D., McKenzie, E.H., Bhat, D.J., Phillips, A.J., Wanasinghe, D.N., Samarakoon, M.C., Jayawardena, R.S., Dissanayake, A.J., Tennakoon, D.S., Doilom, M., Phookamsak, R., Tang, A.M., Xu, J.C., Mortimer, P.E., Promputtha, I., Maharachchikumbura, S.S., Khan, S. & Karunarathna, S.C. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. https://doi.org/10.1007/s13225-018-0408-6
- Wang, S., Kirschner, R., Wang, Y.W. & Hou, C.L. (2014b) Hypohelion from China. Mycological Progress 13: 781–789. https://doi.org/10.1007/s11557-014-0961-7
- White, T.J., Burns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols, a guide to methods and applications. Academic, San Diego, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Zhang, J.F., Liu, J.K., Hyde, K.D., Ekanayaka, A.H. & Liu, Z.Y. (2020) Morpho-phylogenetic evidence reveals new species in Rhytismataceae (Rhytismatales, Leotiomycetes, Ascomycota) from Guizhou Province, China. MycoKeys 76: 81–106. https://doi.org/10.3897/mycokeys.76.58465
- Zoller, S., Scheidegger, C. & Sperisen, C. (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31: 511–516. https://doi.org/10.1006/lich.1999.0220