Abstract
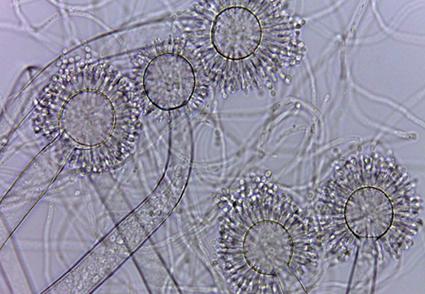
In this study we describe a new species of Aspergillus in section Cremei deposited in the URM culture collection (Micoteca URM Profa. Maria Auxiliadora Cavalcanti) of the Departamento de Micologia, Universidade Federal de Pernambuco (Recife, Pernambuco, Brazil). Another strain was obtained from samples taken from pollen pots inside nests of Melipona scutellaris (stingless bee). Morphological and phylogenetic evidence based on sequencing of ITS rDNA, BenA, CaM and rpb2 markers supported the description of taxonomic novelty. Aspergillus capibaribensis sp. nov. is morphologically similar and phylogenetically close to A. flaschentraegeri - the distinctive features are uniseriate and hyaline conidiophores, short and smooth conidia and smaller vesicles compared to the micromorphological structures of A. flaschentraegeri. This record also expanded the knowledge of species in Aspergillus section Cremei in a tropical country such as Brazil.
References
- Alves, V.C.S., Lira, R.A., Lima, J.M.S., Barbosa, R.N., Bento, D.M., Barbier, E., Bernard, E., Souza-Motta, C.M. & Bezerra, J.D.P. (2022) Unravelling the fungal darkness in a tropical cave: richness and the description of one new genus and six new species. Fungal Systematics and Evolution 10: 139–167. https://doi.org/10.3114/fuse.2022.10.06
- Barbosa, R.N., Bezerra, J.D.P., Santos, A.C.S., Melo, R.F.R., Houbraken, J., Oliveira, N.T. & Souza-Motta, C.M. (2020) Brazilian tropical dry forest (Caatinga) in the spotlight: an overview of species of Aspergillus, Penicillium and Talaromyces (Eurotiales) and the description of P. vascosobrinhous sp. nov. Acta Botanica Brasilica 34: 409–429. https://doi.org/10.1590/0102-33062019abb0411
- Barbosa, R.N., Bezerra, J.D.P., Souza-Motta, C.M., Frisvad, J., Samson, R.A., Oliveira, N.T. & Houbraken, J. (2018) New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie van Leeuwenhoek 111: 1883–1912. https://doi.org/10.1007/s10482-018-1081-1
- Barbosa, R.N., Santos, J.E.F., Bezerra, J.D.P., Istel, U., Houbraken, J., Oliveira, N.T. & Souza-Motta, C.M. (2022) Brazilian Atlantic Forest and Pampa Biomes in the spotlight: an overview of Aspergillus, Penicillium, and Talaromyces (Eurotiales) species and the description of Penicillium nordestinense sp. nov. Acta Botanica Brasilica 36: e2021abb0390. https://doi.org/10.1590/0102-33062021abb0390
- Barros-Correia, A.C.R.B., Barbosa, R.N., Frisvad, J.C., Houbraken, J. & Souza-Motta, C.M. (2020) The polyphasic re-identification of a Brazilian Aspergillus section Terrei collection led to the discovery of two new species. Mycological Progress 19: 885–903. https://doi.org/10.1007/s11557-020-01605-4
- Belfiori, B., Rubini, A. & Riccioni, C. (2021) Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy. Diversity 13: 535. https://doi.org/10.3390/d13110535
- Bennett, S.E.B.J.W. (2007) An overview of the genus Aspergillus. The Aspergilli, pp. 23–34.
- Bezerra, J.D.P., Felipe, M.T., Paiva, L.M., Magalhães, O.M.C., Silva-Nogueira, E.B., Silva, G.A. & Souza-Motta, C.M. (2020) Phylogenetic placement of Tritirachium strains from the URM culture collection originally founded by Augusto Chaves Batista (1916-1967) in Brazil, and the description of T. batistae sp. nov. Acta Botanica Brasilica 34: 290–300. https://doi.org/10.1590/0102-33062019abb0356
- Bovio, E., Garzoli, L., Poli, A., Prigione, V., Firsova, D., McCormack, G.P. & Varese, G.C. (2018) The culturable mycobiota associated with three Atlantic sponges, including two new species: Thelebolus balaustiformis and T. spongiae. Fungal Systematics and Evolution 1: 141–167. https://doi.org/10.3114/fuse.2018.01.07
- de Vries, R.P., Riley, R., Wiebenga, A., Aguilar-Osorio, G., Amillis, S., Uchima, C.A., Anderluh, G., Asadollahi, M., Askin, M., Barry, K., Battaglia, E., Bayram, Ö., Benocci, T., Braus-Stromeyer, S.A., Caldana, C., Cánovas, D., Cerqueira, G.C., Chen, F., Chen, W., Choi, C., Clum, A., Dos Santos, R.A., Damásio, A.R., Diallinas, G., Emri, T., Fekete, E., Flipphi, M., Freyberg, S., Gallo, A., Gournas, C., Habgood, R., Hainaut, M., Harispe, M.L., Henrissat, B., Hildén, K.S., Hope, R., Hossain, A., Karabika, E., Karaffa, L., Karányi, Z., Kraševec, N., Kuo, A., Kusch, H., LaButti, K., Lagendijk, E.L., Lapidus, A., Levasseur, A., Lindquist, E., Lipzen, A., Logrieco, A.F., MacCabe, A., Mäkelä, M.R., Malavazi, I., Melin, P., Meyer, V., Mielnichuk, N., Miskei, M., Molnár, Á.P., Mulé, G., Ngan, C.Y., Orejas, M., Orosz, E., Ouedraogo, J.P., Overkamp, K.M., Park, H.S., Perrone, G., Piumi, F., Punt, P.J., Ram, A.F., Ramón, A., Rauscher, S., Record, E., Riaño-Pachón, D.M., Robert, V., Röhrig, J., Ruller, R., Salamov, A., Salih, N.S., Samson, R.A., Sándor, E., Sanguinetti, M., Schütze, T., Sepčić, K., Shelest, E., Sherlock, G., Sophianopoulou, V., Squina, F.M., Sun, H., Susca, A., Todd, R.B., Tsang, A., Unkles, S.E., van de Wiele, N., van Rossen-Uffink, D., Oliveira, J.V., Vesth, T.C., Visser, J., Yu, J.H., Zhou, M., Andersen, M.R., Archer, D.B., Baker, S.E., Benoit, I., Brakhage, A.A., Braus, G.H., Fischer, R., Frisvad, J.C., Goldman, G.H., Houbraken, J., Oakley, B., Pócsi, I., Scazzocchio, C., Seiboth, B., vanKuyk, P.A., Wortman, J., Dyer, P.S. & Grigoriev, I.V. (2017) Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biology 18 (1): 28. https://doi.org/10.1186/s13059-017-1151-0
- Felipe, M.T., Barbosa, R.N., Bezerra, J.D.P. & Souza-Motta, C.M. (2023) Production of kojic acid by Aspergillus species: trends and applications. Fungal Biology Reviews 45: 100313. https://doi.org/10.1016/j.fbr.2023.100313
- Ferraz, R.E., Lima, P.M., Pereira, D.S., Freitas, C.C.O. & Feijó, F.M.C. (2008) Microbiota fúngica de Melipona subnitida Ducke (Hymenoptera: Apidae). Neotropical Entomology 37: 345–346. https://doi.org/10.1590/S1519-566X2008000300017
- Flannigan, B. (1986) Aspergillus clavatus - an allergenic, toxigenic deteriorgen of cereals and cereal products. International Biodeterioration 22: 79–89.
- Frisvad, J.C., Hubka, V., Ezekiel, C.N., Hong, S.B., Nováková, A., Chen, A.J., Arzanlou, M.., Larsen, TO., Sklenář, F., Mahakarnchanakul, W., Samson, R.A. & Houbraken, J. (2019) Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology 93: 1–63. https://doi.org/10.1016/j.simyco.2018.06.001
- Gams, W., Christensen, M., Onions, A.H., Pitt, J.I. & Samson, R.A. (1986) Infrageneric Taxa of Aspergillus. In: Samson, R.A. & Pitt, J.I. (Eds.) Advances in Penicillium and Aspergillus Systematics. Plenum Press, New York, pp. 55–62. https://doi.org/10.1007/978-1-4757-1856-0_5
- Glaser, M. (2003) Interrelations between mangrove ecosystem, local economy and social sustainability in Caete Estuary, North Brazil. Wetlands Ecology and Management 11: 265–272. https://doi.org/10.1023/A:1025015600125
- Glass, N.L. & Donaldson, G.C. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. https://doi.org/10.1128/aem.61.4.1323-1330.1995
- Glässnerová, K., Sklenář, F., Jurjević, Ž., Houbraken, J., Yaguchi, T., Visagie, C.M., Gené, J., Siqueira, J.P.Z., Kubátová, A., Kolařík, M. & Hubka, V. (2022) A monograph of Aspergillus section Candidi. Studies in Mycology 102: 1–51. https://doi.org/10.3114/sim.2022.102.01
- Halsey, C., Lumley, H. & Luckit, J. (2010) Necrotising external otitis caused by Aspergillus wentii: a case report. Mycoses 54: 211–213. https://doi.org/10.1111/j.1439-0507.2009.01815.x
- Hong, S.B. (2013) Aspergillus luchuensis, an industrially important black Aspergillus in East Asia. Plos One 8: 1–9. https://doi.org/10.1371/journal.pone.0063769
- Houbraken, J., Kocsubé, S., Visagie, C.M., Yilmaz, N., Wang, X.C., Meijer, M., Kraak, B., Hubka, V., Bensch, K., Samson, R.A. & Frisvad, J.C. (2020) Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. https://doi.org/10.1016/j.simyco.2020.05.002
- Hubka, V., Kubatova, A., Mallatova, N., Sedlacek, P., Melichar, J., Skorepova, M., Mencl, K., Lyskova, P., Sramkova, B., Chudickova, M., Hamal, P. & Kolarik, M. (2012) Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Medical Mycology 50: 601–610. https://doi.org/10.3109/13693786.2012.667578
- Hubka, V., Nováková, A., Samsom, R.A., Houbraken, J., Frisvad, J.C., Sklenár, F., Varga, J. & Kolařík, M. (2016) Aspergillus europaeus sp. nov., a widely distributed soil-borne species related to A. wentii (section Cremei). Plant Systematics and Evolution 302: 641–650. https://doi.org/10.1007/s00606-016-1293-7
- Katoh, K.L. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780.
- Kinoshita, K. (1932) Uber die Production von Itaconsaure and Mannit durch einen neuen Schimmelpiz Aspergillus itacnicus. Acta Phytochim 5: 271–287.
- Kocsubé, S., Perrone, G., Magistà, D., Houbraken, J., Varga, J., Szigeti, G., Hubka, V., Hong, S.B., Frisvad, J.C. & Samson, R.A. (2016) Aspergillus is monophyletic: Evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology 85: 199–213. https://doi.org/10.1016/j.simyco.2016.11.006
- Kumar, A. (2020) Aspergillus nidulans: A Potential Resource of the Production of the Native and Heterologous Enzymes for Industrial Applications. International Journal of Microbiology 2020: 1–11. https://doi.org/10.1155/2020/8894215
- Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
- Lee, J.W., Lee, W., Perera, R.H. & Lim, Y.W. (2023) Long-Term Investigation of Marine-Derived Aspergillus Diversity in the Republic of Korea. Mycobiology 51 (6): 436–444. https://doi.org/10.1080/12298093.2023.2279342
- LeMense, E.H., Corman, J., Van Lanen, J.M. & Langlykke, A.F. (1947) Production of mold amylases in submerged culture. Journal of Bacteriology 54: 149–159.
- Liu, Y.J., Whelen, S. & Hall, B. (1999) Phylogenetic relationship among Ascomycetes: Evidence from an RNA Polymerase II Subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Maddison, W.P. & Maddison, D.R. (2019) Mesquite: A modular system for evolutionary analysis. Version 3.61. Available from: http://mesquiteproject.org (accessed 17 October 2022)
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Stewart, C. (Eds.) Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, pp. 1–8.
- Mituishi, M.P., Menezes, L.H.S., Tavares, I.M.C., Freitas, J.S., Souza-Motta, C.M., Bezerra, J.L., Costa, A.M., Uetanabaro, A.P.T., Franco, M. & Oliveira, J.R. (2022) Potential of Aspergillus niger Tiegh 8285 in the bioremediation of water contaminated with benzonitrile. Research, Society and Development 11: e42711831078. https://doi.org/10.33448/rsd-v11i8.31078
- Nguyen, T.T.T., Pangging, M., Bangash, N.K. & Lee, H.B. (2020) Five New Records of the Family Aspergillaceae in Korea, Aspergillus europaeus, A. pragensis, A. tennesseensis, Penicillium fluviserpens, and P. scabrosum. Mycobiology 48 (2): 81–94. https://doi.org/10.1080/12298093.2020.1726563
- Peterson, S.W. (2008) Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. https://doi.org/10.3852/mycologia.100.2.205
- Posada, D. (2008) jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256.
- Rambaut, A. (2016) FigTree, version 1.4.3. Edinburgh, Institute of Evolutionary Biology, University of Edinburgh.
- Raper, K.B. & Fennel, D.I. (1965) The Genus Aspergillus. The Williams and Wilkins. Baltimore, Maryland, pp. 686.
- Raper, K.B. & Fennell, D.I. (1969) The genus Aspergillus. The Williams and Wilkins. Baltimore, Maryland, pp. 686.
- Rayner, R.W. (1970) A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society. Kew, Surrey, UK, pp 36.
- Rintoul, T.L., Eggertson, Q.A. & Lévesque, C.A. (2012) Multigene phylogenetic analyses to delimit new species in fungal plant pathogens. Methods in Molecular Biology 835: 549–569.
- Roehr, M., Kubicek, C.P. & Kominek, J. (1992) Industrial acids and other small molecules. In: Bennett, J.W. & Klich, M.A. (Eds.) Aspergillus: Biology and industrial applications. Butterworth-Heinemann, Boston, pp. 91131.
- Ronquist, F., Teslenko, M., Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
- Samson, R.A., Visagie, C.M., Houbraken, J., Hong, S.B., Hubka, V., Klaassen, C.H.W., Perrone, G., Seifert, K.A., Susca, A., Tanney, J.B., Varga, J., Kocsubé, S., Szigeti, G., Yaguchi, T. & Frisvad, J.C. (2014) Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology 78: 141–173. https://doi.org/10.1016/j.simyco.2014.07.004
- Shearer, C.A., Descals, E., Kohlmeyer, B., Kohlmeyer, J., Marvanova, L., Padgett, D., Porter, D., Raja, H.A., Schmit, J.P., Thorton, H.A. & Voglymayr, H. (2007) Fungal diversity in aquatic habitats. Biodiversity and Conservation 16: 49–67. https://doi.org/10.1007/s10531-006-9120-z
- Silva, J.J., Fungaro, M.H.P., Wang, X., Larsen, T.O., Frisvad, J.C., Taniwaki, M.H. & Iamanaka, B.T. (2022) Deep Genotypic Species Delimitation of Aspergillus Section Flavi Isolated from Brazilian Foodstuffs and the Description of Aspergillus annui sp. nov. and Aspergillus saccharicola sp. nov. Journal of Fungi 8: 1279. https://doi.org/10.3390/jof8121279
- Souza-Motta, C.M. & Barbosa, R.N. (2022) Micoteca URM da UFPE: um importante repositório da micodiversidade brasileira. Boletim Micobiota 2 (4).
- Spadoni, A., Ippolito, A., Admane, N. & Sanzani, S.M. (2020) First report of Aspergillus europaeus causing postharvest bulb rot of garlic in Italy. Journal of Plant Pathology 102: 601. https://doi.org/10.1007/s42161-019-00485-2
- Stamatakis, A. (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 13121313.
- Steiger, M.G., Blumhoff, M.L., Mattanovich, D. & Sauer, M. (2013) Biochemistry of microbial itaconic acid production. Frontiers in Microbiology 4: 1–5. https://doi.org/10.3389/fmicb.2013.00023
- Sung, G.H., Sung, J.M., Hywel-Jones, N.L. & Spatafora, J.W. (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44: 1204–1223. https://doi.org/10.1016/j.ympev.2007.03.011
- Thatoi, H., Behera, B.C., Mishra, R.R. & Dutta, S.K. (2013) Biodiversity and biotechnological potential of microorganisms from mangrove ecosystems: a review. Annals of Microbiology 63: 1–19. https://doi.org/10.1007/s13213-012-0442-7
- Thom, C. & Church, M.B. (1926) The Aspergillus. Baltimore: Williams & Wilkins.
- Visagie, C.M., Yilmaz, N., Kocsubé, S., Frisvad, J.C., Hubka, V., Samson, R.A. & Houbraken, J. (2024) A review of recently introduced Aspergillus, Penicillium, Talaromyces and other Eurotiales species. Studies in Mycology 107: 1–66. https://doi.org/10.3114/sim.2024.107.01
- Wells, J.M., Cole, R.J. & Kirksey, J.W. (1975) Emodin, a toxic metabolite of Aspergillus wentii isolated from weevil-damaged chestnuts. Applied Microbiology 30: 26.
- White, T.J., Bruns, T. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylognetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, J.W (Eds.) A Guide to Molecular Methods and 76 Applications. Academic Press, New York, pp. 315–322. http://dx.doi.org/10.1016/B978-0-12-372180-8.50042-1