Abstract
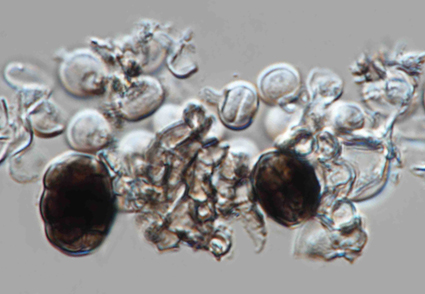
During investigations of hyphomycetous fungi from medicinal plants in Yunnan Province, China, a novel species of Hermatomyces was discovered. Hermatomyces pyriformis is introduced based on morphology and phylogenetic analyses of combined LSU, ITS, tef1-α and rpb2 sequence data. This fungus is characterized by having sporodochial conidiomata and determinate, hyaline, pyriform conidiogenous cells bearing brown, globose to subglobose or ellipsoidal, muriform lenticular conidia with dark brown center and brown peripheral cells. Phylogenetic results indicated that two isolates of H. pyriformis represent a distinct lineage and can be recognized as a new species. A detailed description and illustration are provided for the new species.
References
- Abtahi, F. & Nourani, S.L. (2017) The most important fungal diseases associated with some useful medicinal plants. In: Ghorbanpour, M. & Varma, A. (Eds.) Medicinal Plants and Environmental Challenges. Springer, Cham, pp. 279–293. https://doi.org/10.1007/978-3-319-68717-9_16
- Ali, S.I., Sheikh, W.M., Rather, M.A., Venkatesalu, V., Muzamil Bashir, S. & Nabi, S.U. (2021) Medicinal plants: treasure for antiviral drug discovery. Phytotherapy Research : PTR 35 (7): 3447–3483. https://doi.org/10.1002/ptr.7039
- Ariyawansa, H.A., Hyde, K.D., Jayasiri, S.C., Buyck, B., Chethana, K.W.T., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Jayawardena, R.S., Lücking, R., Ghobad-Nejhad, M., Niskanen, T., Thambugala, K.M., Spirin, V. & Hernawati (2015) Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75: 27–274. https://doi.org/10.1007/s13225-015-0346-5
- Atanasov, A.G., Zotchev, S.B., Dirsch, V.M., Orhan, I.E., Banach, M., Rollinger, J.M., Barreca, D., Stadler, M., Daglia, M., Verpoorte, R. & Supuran, C.T. (2021) Natural products in drug discovery: advances and opportunities. Nature Reviews Drug Discovery 20 (3): 200–216. https://doi.org/10.1038/s41573-020-00114-z
- Bhagat, J., Kaur, A., Sharma, M., Saxena, A.K. & Chadha, B.S. (2012) Molecular and functional characterization of endophytic fungi from traditional medicinal plants. World Journal of Microbiology & Biotechnology 28: 963–971. https://doi.org/10.1007/s11274-011-0894-0
- Bitancourt, A.A. & Jenkins, A.E. (1950) Estudos sôbre as Miriangiales. I. Dez novas espécies de Elsinoaceas descobertas no Brasil. Arquivos do Instituto Biológico de São Paulo 19: 93–109.
- Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15): 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
- Castañeda-Ruiz, R. & Heredia, G. (2000) Two new dematiaceous hyphomycetes on Cyathea from Mexico. Cryptogamie Mycologie 21: 221–228. https://doi.org/10.1016/S0181-1584(00)01047-2
- Cole, I.B., Saxena, P.K. & Murch, S.J. (2007) Medicinal biotechnology in the genus Scutellaria. In Vitro Cellular & Developmental Biology–Plant 43: 318–327. https://doi.org/10.1007/s11627-007-9055-4
- de Silva, N.I., Hyde, K.D., Lumyong, S., Phillips, A.J.L., Bhat, D.J., Maharachchikumbura, S.S.N., Thambugala, K.M., Tennakoon, D.S., Suwannarach, N. & Karunarathna, S.C. (2022) Morphology, phylogeny, host association and geography of fungi associated with plants of Annonaceae, Apocynaceae and Magnoliaceae. Mycosphere 13 (1): 955–1076. https://doi.org/10.5943/mycosphere/13/1/12
- Delgado, G., Koukol, O., Heredia, G. & Piepenbring, M. (2020) Texas microfungi: Hermatomyces amphisporus (Pleosporales, Dothideomycetes) revisited. Czech Mycology 72 (1): 95–107. https://doi.org/10.33585/cmy.72107
- Dissanayake, A.J., Bhunjun, C.S., Maharachchikumbura, S.S.N. & Liu, J.K. (2020) Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11 (1): 2652–2676. https://doi.org/10.5943/mycosphere/11/1/18
- Doilom, M., Dissanayake, A.J., Wanasinghe, D.N., Boonmee, S., Liu, J.K., Bhat, D.J., Taylor, J.E., Bahkali, A.H., McKenzie, E.H.C. & Hyde, K.D. (2016) Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Diversity 82: 107–182. https://doi.org/10.1007/s13225-016-0368-7
- Du, T.Y., Karunarathna, S.C., Zhang, X., Dai, D.Q., Mapook, A., Suwannarach, N., Xu, J.C., Stephenson, S.L., Elgorban, A.M., Al-Rejaie, S. & Tibpromma, S. (2022) Endophytic fungi associated with Aquilaria sinensis (agarwood) from China show antagonism against bacterial and fungal pathogens. Journal of Fungi 8 (11): 1197. https://doi.org/10.3390/jof8111197
- Hashimoto, A., Matsumura, M., Hirayama, K., Yonezawa, H. & Tanaka, K. (2016) Taxonomy and phylogeny of Cryptocoryneum (Pleosporales, Dothideomycetes). Mycological Progress 15: 1–12. https://doi.org/10.1007/s11557-016-1186-8
- Hashimoto, A., Matsumura, M., Hirayama, K. & Tanaka, K. (2017) Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Persoonia 39: 51–73. https://doi.org/10.3767/persoonia.2017.39.03
- Hassan, B.A. (2012) Medicinal plants (importance and uses). Pharmaceutica Analytica Acta 3: 10. https://doi.org/10.4172/2153-2435.1000e139
- Hawksworth, D.L. (2011) Ascospore sculpturing and generic concepts in the Testudinaceae (syn. Zopfiaceae). Canadian Journal of Botany 57: 91–99. https://doi.org/10.1139/b79-017
- Helaly, S.E., Thongbai, B. & Stadler, M. (2018) Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Natural Product Reports 35 (9): 992–1014. https://doi.org/10.1039/c8np00010g
- Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H.C., Sarma, V.V., Lücking, R., Boonmee, S., Bhat, J.D., Liu, N.G., Feng, Y. & Xie, N. (2020) Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105: 17–318. https://doi.org/10.1007/s13225-020-00462-6
- Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., McKenzie, E.H.C., Jones, E.B.G., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhou, J.L. & Zhu, L. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x
- Hyde, K.D., Norphanphoun, C., Abreu, V.P., Bazzicalupo, A., Thilini Chethana, K.W., Clericuzio, M., Dayarathne, M.C., Dissanayake, A.J., Ekanayaka, A.H., Zhao, R.L. & Mortimer, P.E. (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87: 1–235. https://doi.org/10.1007/s13225-017-0391-3
- Hyde, K., Tennakoon, D., Jeewon, R., Bhat, D.J., Maharachchikumbura, S., Rossi, W., Leonardi, M., Lee, H., Mun, H.Y., Houbraken, J., Nguyễn, T., Jeon, S., Frisvad, J., Wanasinghe, D., Lücking, R., Aptroot, A., Cáceres, M., Karunarathna, S., Hongsanan, S. & Doilom, M. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. https://doi.org/10.1007/s13225-019-00429-2
- Hyde, K.D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D.J., Jones, E.B.G., Liu, N.G., Abeywickrama, P.D., Mapook, A., Wei, D., Lumyong, S., Xu, J.C. & Sheng, J. (2020a) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. https://doi.org/10.1007/s13225-020-00439-5
- Hyde, K.D., Jeewon, R., Chen, Y.J., Bhunjun, C.S., Calabon, M.S., Jiang, H.B., Lin, C.G., Norphanphoun, C., Sysouphanthong, P., Pem, D., Tibpromma, S., Zhang, Q., Doilom, M., Jayawardena, R.S., Liu, J.K., Maharachchikumbura, S.S.N., Phukhamsakda, C., Phookamsak, R., Al-Sadi, A.M., Thongklang, N., Wang, Y., Gafforov, Y., Gareth Jones, E.B. & Lumyong, S. (2020b) The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 103: 219–271. https://doi.org/10.1007/s13225-020-00458-2
- Jayasiri, S.C., Jones, E.B.G., Kang, J.C., Promputtha, I., Bahkali, A.H. & Hyde, K.D. (2016) A new species of genus Anteaglonium (Anteagloniaceae, Pleosporales) with its asexual morph. Phytotaxa 263 (3): 233–244. https://doi.org/10.11646/phytotaxa.263.3.4
- Jenkins, A.E. (1932) Elsinoë on apple and pear. Journal of Agricultural Research 44: 689–700.
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30 (4): 772–780. https://doi.org/10.1093/molbev/mst010
- Keshri, P., Rai, N., Verma, A., Kamble, S., Barik, S., Mishra, P., Singh, S., Salvi, P. & Gautam, V. (2021) Biological potential of bioactive metabolites derived from fungal endophytes associated with medicinal plants. Mycological Progress 20: 577–594. https://doi.org/10.1007/s11557-021-01695-8
- Kohlmeyer, J. & Volkmann-Kohlmeyer, B. (1990) Revision of marine species of Didymosphaeria (Ascomycotina). Mycological Research 94 (5): 685–690. https://doi.org/10.1016/S0953-7562(09)80669-2
- Koukol, O., Delgado, G., Hofmann, T.A. & Piepenbring, M. (2018) Panama, a hot spot for Hermatomyces (Hermatomycetaceae, Pleosporales) with five new species, and a critical synopsis of the genus. IMA Fungus 9 (1): 107–141. https://doi.org/10.5598/imafungus.2018.09.01.08
- Koukol, O. & Delgado, G. (2019) Do not forget Africa-revision of fungarium collections at Kew revealed a new species of Hermatomyces (Hermatomycetaceae, Pleosporales). Nova Hedwigia 109 (3/4): 413–423. https://doi.org/10.1127/nova_hedwigia/2019/0559
- Larget, B. & Simon, D.L. (1999) Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution 16 (6): 750–759. https://doi.org/10.1093/oxfordjournals.molbev.a026160
- Larsson, A. (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278. https://doi.org/10.1093/bioinformatics/btu531
- Leão-Ferreira, S., Gusmao, L. & Castañeda-Ruiz, R. (2013) Conidial fungi from the semi-arid Caatinga biome of Brazil. Three new species and new records. Nova Hedwigia 96: 479–494. https://doi.org/10.1127/0029-5035/2013/0084
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16 (12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Liu, J.K., Chomnunti, P., Cai, L., Phookamsak, R., Chukeatirote, R., Jones, E.B.G., Moslem, M. & Hyde, K.D. (2010) Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia 62 (2): 261–276.
- Locquin, M. (1984) Mycologie générale et structurale: 202. Masson, Paris.
- Ma, X.Y., Maharachchikumbura, S.S.N., Chen, B.W., Hyde, K.D., McKenzie, E.H.C., Chomnunti, P. & Kang, J.C. (2019) Endophytic pestalotiod taxa in Dendrobium orchids. Phytotaxa 419: 268–286. https://doi.org/10.11646/phytotaxa.419.3.2
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE) 14: 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Mugambi, G.K. & Huhndorf, S.M. (2009) Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7: 453–464. https://doi.org/10.1017/S147720000999020X
- Müller, E. & von Arx, J.A. (1962) Die Gattungen der didymosporen Pyrenomyceten, vol. 11. Büchler.
- Nalawade, S., Sagare, A., Lee, C.Y., Kao, C.L. & Tsay, H.S. (2003) Studies on tissue culture of Chinese medicinal plant resources in Taiwan and their sustainable utilization. Botanical Bulletin-Academia Sinica Taipei 44 (2): 79–98.
- Niu, X.P., Gao, H., Qi, J.M., Chen, M.C., Tao, A.F., Xu, J.T., Dai, Z.G. & Su, J.G. (2016) Colletotrichum species associated with jute (Corchorus capsularis L.) anthracnose in southeastern China. Scientific Reports 6 (1): 25179. https://doi.org/10.1038/srep25179
- Nuankaew, S., Suetrong, S., Wutikhun, T. & Pinruan, U. (2019) Hermatomyces trangensis sp. nov., a new dematiaceous hyphomycete (Hermatomycetaceae, Pleosporales) on sugar palm in Thailand. Phytotaxa 391 (5): 277–288. https://doi.org/10.11646/phytotaxa.391.5.1
- Nylander, J. (2008) MrModeltest 2 v. 2.3 (Program for selecting DNA substitution models using PAUP*). Evolutionary Biology Centre, Uppsala, Sweden.
- Phookamsak, R., Norphanphoun, C., Tanaka, K., Dai, D.Q., Luo, Z.L., Liu, J.K., Su, H.Y., Bhat, D.J., Bahkali, A.H., Mortimer, P.E., Xu, J.C. & Hyde, K.D. (2015) Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Diversity 74: 143–197. https://doi.org/10.1007/s13225-015-0352-7
- Phukhamsakda, C., McKenzie, E.H.C., Phillips, A.J.L., Gareth Jones, E.B., Jayarama Bhat, D., Stadler, M., Bhunjun, C.S., Wanasinghe, D.N., Thongbai, B., Camporesi, E., Ertz, D., Jayawardena, R.S., Perera, R.H., Ekanayake, A.H., Tibpromma, S., Doilom, M., Xu, J.C. & Hyde, K.D. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102: 1–203. https://doi.org/10.1007/s13225-020-00448-4
- Rahman, I.U., Afzal, A., Iqbal, Z., Ijaz, F., Ali, N., Shah, M., Ullah, S. & Bussmann, R.W. (2019) Historical perspectives of ethnobotany. Clinics in Dermatology 37 (4): 382–388. https://doi.org/10.1016/j.clindermatol.2018.03.018
- Rannala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10.1007/BF02338839
- Rao, V. & de Hoog, G.S. (1986) New or critical hyphomycetes from India. Studies in Mycology 28: 1–84.
- Rasool, A., Bhat, K.M., Sheikh, A.A., Jan, A. & Hassan, S. (2020) Medicinal plants: role, distribution and future. Journal of Pharmacognosy and Phytochemistry 9 (2): 2111–2114.
- Rehner, S.A. & Buckley, E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97 (1): 84–98. https://doi.org/10.1080/15572536.2006.11832842
- Ren, G.C., Wanasinghe, D.N., Monkai, J., Mortimer, P.E., Hyde, K.D., Xu, J.C., Pang, A. & Gui, H. (2021) Novel saprobic Hermatomyces species (Hermatomycetaceae, Pleosporales) from China (Yunnan Province) and Thailand. MycoKeys 82: 57–79. https://doi.org/10.3897/mycokeys.82.67973
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
- Samy, R.P. & Gopalakrishnakone, P. (2007) Current status of herbal and their future perspectives. Nature Precedings. https://doi.org/10.1038/npre.2007.1176.1
- Schmidt, B.M. (2017) Ethnobotany. In: Schmidt, B.M. & Klaser Cheng, D.M. (Eds.) Ethnobotany: a phytochemical perspective. pp. 1–109. https://doi.org/10.1002/9781118961933.ch1
- Senanayake, I.C., Rathnayaka, A.R., Sandamali, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11 (1): 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
- Spegazzini, C.L. (1911) Mycetes argentinenses. Series V. Anales Museo Nacional de Historia Natural Buenos Aires 3: 446.
- Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 (21): 2688–2690. https://doi.org/10.1093/bioinformatics/btl446
- Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57 (5): 758–771. https://doi.org/10.1080/10635150802429642
- Sun, Y.R., Jayawardena, R.S., Sun, J.E. & Wang, Y. (2023) Pestalotioid species associated with medicinal plants in Southwest China and Thailand. Microbiology Spectrum 11 (1): e0398722. https://doi.org/10.1128/spectrum.03987-22
- Tanaka, K., Hirayama, K., Yonezawa, H., Hatakeyama, S., Harada, Y., Sano, T., Shirouzu, T. & Hosoya, T. (2009) Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Studies in Mycology 64 (2): 175–209. https://doi.org/10.3114/sim.2009.64.10
- Tennakoon, D.S., Kuo, C.H., Maharachchikumbura, S.S.N., Thambugala, K.M., Gentekaki, E., Phillips, A.J.L., Bhat, D.J., Wanasinghe, D.N., de Silva, N.I., Promputtha, I. & Hyde, K.D. (2021) Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Diversity 108: 1–215. https://doi.org/10.1007/s13225-021-00474-w
- Tibpromma, S., Bhat, D.J., Doilom, M., Lumyong, S., Nontachaiyapoom, S., Yang, J.B. & Hyde, K.D. (2016) Three new Hermatomyces species (Lophiotremataceae) on Pandanus odorifer from Southern Thailand. Phytotaxa 275 (2): 127–139. https://doi.org/10.11646/phytotaxa.275.2.4
- Tibpromma, S., Hyde, K.D., Jeewon, R., Maharachchikumbura, S.S.N., Liu, J.K., Bhat, D.J., Jones, E.B.G., McKenzie, E.H.C., Nontachaiyapoom, S., Wen, T.C. & Karunarathna, S.C. (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83: 1–261. https://doi.org/10.1007/s13225-017-0378-0
- Tibpromma, S., Hyde, K.D., McKenzie, E.H.C., Bhat, D.J., Phillips, A.J.L., Wanasinghe, D.N., Samarakoon, M.C., Jayawardena, R.S., Dissanayake, A.J., Tennakoon, D.S., Doilom, M., Phookamsak, R., Tang, A.M.C., Xu, J.C., Mortimer, P.E., Promputtha, I., Maharachchikumbura, S.S.N., Khan, S. & Karunarathna, S.C. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. https://doi.org/10.1007/s13225-018-0408-6
- Vaidya, G., Lohman, D.J. & Meier, R. (2011) SequenceMatrix concatenation software for the fast assembly of multi gene datasets with character set and codon information. Cladistics 27 (2): 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172 (8): 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18 (1): 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1
- Wijayawardene, D.N.N., McKenzie, E.H.C. & Hyde, K.D. (2012) Towards incorporating anamorphic fungi in a natural classification–Checklist and notes for 2011. Mycosphere 3 (2): 157–228. https://doi.org/10.5943/mycosphere/3/2/5
- Wijayawardene, N.N., Phillips, A.J.L., Tibpromma, S., Dai, D.Q., Selbmann, L., Monteiro, J.S., Aptroot, A., Flakus, A., Rajeshkumar, K.C., Coleine, C., Pereira, D.S., Fan, X., Zhang, L., Maharachchikumbura, S.S.N., Souza, M.F., Kukwa, M., Suwannarach, N., Rodriguez-Flakus, P., Ashtekar, N., Dauner, L., Tang, L.Z., Jin, X.C. & Karunarathna, S.C. (2021) Looking for the undiscovered asexual taxa: case studies from lesser studied life modes and habitats. Mycosphere 12 (1): 1186–1229. https://doi.org/10.5943/mycosphere/12/1/17
- Wu, N., Thilini Chethana, K.W. & Liu, J.K. (2024) Velutinus, a novel genus in Patellariopsidaceae from medicinal plants in Sichuan Province, China. Phytotaxa 638 (2): 165–174. https://doi.org/10.11646/phytotaxa.638.2.5
- Yang, J., Liu, L., Jones, E., Hyde, K., Liu, Z.Y., Bao, D.F., Liu, N.G., Li, W., Shen, H., Yu, X.D. & Liu, J.K. (2023) Freshwater fungi from karst landscapes in China and Thailand. Fungal Diversity 119: 1–212. https://doi.org/10.1007/s13225-023-00514-7
- Zhang, Y., Schoch, C.L., Fournier, J., Crous, P.W., de Gruyter, J., Woudenberg, J.H.C., Hirayama, K., Tanaka, K., Pointing, S.B., Spatafora, J.W. & Hyde, K.D. (2009) Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85–102. https://doi.org/10.3114/sim.2009.64.04
- Zhang, H., Hyde, K.D., McKenzie, E.H.C., Bahkali, A.H. & Zhou, D.Q. (2012) Sequence data reveals phylogenetic affinities of Acrocalymma aquatica sp. nov., Aquasubmersa mircensis gen. et sp. nov. and Clohesyomyces aquaticus (freshwater coelomycetes). Cryptogamie Mycologie 33 (3): 333–346. https://doi.org/10.7872/crym.v33.iss3.2012.333
- Zhang, Y.Z., Chen, Q., Ma, J., Lu, Y.Z., Chen, H. & Liu, N.G. (2023) Morphological and multi-gene phylogenetic analyses reveal five new hyphomycetes from freshwater habitats. Frontiers in Microbiology 14: 1253239. https://doi.org/10.3389/fmicb.2023.1253239