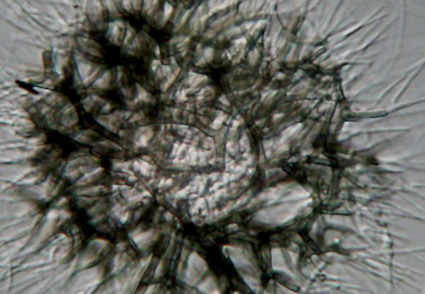
Abstract
We describe Myxotrichum persicum sp. nov. as a seed endophyte of Aegilops triuncialis in Lorestan province of Iran, using morphological traits and sequences of the internal transcribed spacer regions 1 and 2 including the intervening 5.8S nuclear ribosomal DNA (ITS) and partial nuclear 28S ribosomal DNA (LSU) regions. Myxotrichum persicum is differentiated from its closest relative, M. deflexum, by possessing smaller ascomata bearing much shorter, usually unbranched appendages, and smaller, ellipsoidal to fusiform, smooth-walled ascospores. Based on morphology and the multilocus phylogeny of ITS, LSU, MCM7 and RPB1, we herein propose to combine two species of Malbranchea, namely M. circinata and M. flavorosea, into the genus Myxotrichum as Myxotrichum circinatum and M. flavoroseum, respectively.
References
- Addy, H.D., Piercey, M.M. & Currah, R.S. (2005) Micro fungal endophytes in roots. Canadian Journal of Botany 83: 1–13. https://doi.org/10.1139/b04-171
- Arx, J.A. von. (1971) On Arachniotus and related genera of the Gymnoascaceae. Persoonia 6: 371–380.
- Barron, G.L. (1962) New species and new records of Oidiodendron. Canadian Journal of Botany 40: 589–607. https://doi.org/10.1139/b62-055
- Braun, U., Crous, P.W., Dugan, F.M., Groenewald, J.Z. & de Hoog, G.S. (2003) Phylogeny and taxonomy of cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycological Progress 2: 3–18. https://doi.org/10.1007/s11557-006-0039-2
- Calduch, M., Gené, J., Cano, J., Stchigel, A.M. & Guarro, J. (2004) Three new species of Oidiodendron Robak from Spain. Studies in Mycology 50: 159–170.
- Calduch, M., Gené, J., Cano, J. & Guarro, J. (2002) A new species of Oidiodendron with gymnothecium-like sporodochia. Studies in Mycology 47: 211–216.
- Couture, M., Fortin, J.A. & Dalpe, Y. (1983) Oidiodendron griseum Robak: an endophyte of ericoid mycorrhiza in Vaccinum spp. New Phytologist 95: 375–380. https://doi.org/10.1111/j.1469-8137.1983.tb03505.x
- Crous, P.W., Braun, U., Schubert, K. & Groenewald, J.Z. (2007) Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33–56. https://doi.org/10.3114/sim.2007.58.02
- Crous, P.W., Wingfield, M.J., Burgess, T.I., Hardy, G.E., Gené, J., Guarro, J., Baseia, I.G., García, D., Gusmão, L.F.P., Souza-Motta, C.M., Thangavel, R., Adamčík, S., Barili, A., Barnes, C.W., Bezerra, J.D.P., Bordallo, J.J., Cano-Lira, J.F., de Oliveira, R.J.V., Ercole, E., Hubka, V., Iturrieta-González, I., Kubátová, A., Martín, M.P., Moreau, P.A., Morte, A., Ordoñez, M.E., Rodríguez, A., Stchigel, A.M., Vizzini, A., Adbollahzadeh, J., Abreu, V.P., Adamčíková, K., Albuquerque, G.M.R., Alexandrova, A.V., Alvarez, D.E., Armstrong-Cho, C., Banniza, S., Barbosa, R.N., Bellanger, J.M., Bezerra, J.L., Cabral, T.S., Carboň, M., Caicedo, E., Cantillo, T., Carnegie, A.J., Carmo, L.T., Castañeda-Ruiz, R.F., Clement, C.R., Čmoková, A., Conceição, L.B., Cruz, R.H.S.F., Damm, U., da Silva, B.D.B., da Silva, G.A., da Silva, R.M.F., de Santiago, A.L.C.M., de Oliveira, L.F., de Souza, C.A.F., Déniel, F., Dima, B., Dong, G., Edwards, J., Félix, C.R., Fournier, J., Gibertoni, T.B., Hosaka, K., Iturriaga, T., Jadan, M., Jany, J.L., Jurjević, Ž., Kolařík, M., Kušan, I., Landekk, M.F., Leite Cordeiro, T.R., Lima, D.X., Loizides, M., Luo, S., Machado, A.R., Madrid, H., Magalhães, O.M.C., Marinho, P., Matočec, N., Mešić, A., Miller, A.N., Morozova, O.V., Neves, R.P., Nonaka, K., Nováková, A., Oberlies, N.H., Oliveira-Filho, J.R.C., Oliveira, T.G.L., Pap, V., Pereira, O.L., Perrone, G., Peterson, S.W., Pham, T.H.G., Raja, H.A., Raudabaugh, D.B., iŘehulka, J., RodrÃgues-Andrade, E., Saba, M., SchauflerovÃi, A., Shivas, R.G., Simonini, G., Siqueira, J.P.Z., Sousa, J.O., Stajsic, V., Svetasheva, T., Tan, Y.P., Tkalčec, Z., Ullah, S., Valente, P., Valenzuela-Lopez, N., Abrinbana, M., Viana Maques, D.A., Wong, P.T.W., Xavier de Lima, V. & Groenewald, J.Z. (2018) Fungal Planet description sheets: 716–784. Persoonia 40: 240–393. https://doi.org/10.3767/persoonia.2018.40.10
- Currah, R.S. (1985) Taxonomy of the Onygenales: Arthrodermataceae, Gymnoascaceae, Myxotrichaceae and Onygenaceae. Mycotaxon 24: 1–216.
- Currah, R.S. (1994) Peridial morphology and evolution in the prototunicate Ascomycetes. In: Hawksworth, D.L. (ed.) Ascomycete systematics: problems and perspectives in the Nineties. New York: Plenum Press. pp. 281–293. https://doi.org/10.1007/978-1-4757-9290-4_25
- Domsch, K.H., Gams, W. & Anderson T-H. (1980) Compendium of Soil Fungi. Academic Press, U.K.
- Ekanayaka, A.H., Hyde, K.D., Gentekaki, E., McKenzie, E.H.C., Zhao, Q., Bulgakov, T.S. & Camporesi, E. (2019) Preliminary classification of Leotiomycetes. Mycosphere 10: 310–489. https://doi.org/10.5943/mycosphere/10/1/7
- Florea, S., Schardl, C.L. & Hollin, W. (2015) Detection and isolation of Epichloë species, fungal endophytes of grasses. Current Protocols in Microbiology 38: 19A.11.11–19A.11.24. https://doi.org/10.1002/9780471729259.mc19a01s38
- Hambleton, S., Egger, K.N. & Currah, R.S. (1998) The genus Oidiodendron: species delimitation and phylogenetic relationships based on nuclear ribosomal DNA analysis. Mycologia 90: 854–869. https://doi.org/10.2307/3761327
- Hambleton, S. & Currah, R.S. (1997) Fungal endophytes from the roots of alpine and boreal Ericaceae. Canadian Journal of Botany 75: 1570–1581. https://doi.org/10.1139/b97-869
- Hirose, D., Tokiwa, T. & Yaguchi, T. (2022) Two novel Oidiodendron species isolated from roots of Vaccinium boninense in the Bonin Islands, Japan. Phytotaxa 566: 89–104. https://doi.org/10.11646/phytotaxa.566.1.5
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Joseph, B. & Priya, R.M. (2011) Bioactive compounds from endophytes and their potential in pharmaceutical effect: a review. American Journal of Biochemistry and Molecular Biology 1: 291–309. https://doi.org/10.3923/ajbmb.2011.291.309
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
- Kharwar, R.N., Mishra, A., Sharma, V.K., Gond, S.K., Verma, S.K., Kumar, A., Kumar, J., Singh, D.K. & Goutam, J. (2014) Diversity and biopotential of endophytic fungal flora isolated from eight medicinal plants of Uttar Pradesh, India. In: Kharwar, R., Upadhyay, R., Dubey, N. & Raghuwanshi, R. (Eds.) Microbial Diversity and Biotechnology in Food Security. Springer, New Delhi, pp. 23–39. https://doi.org/10.1007/978-81-322-1801-2_3
- Kirk, P.M., Cannon, P.F., Minter, D.W. & Stalpers, J.A. (2008) Ainsworth & Bisby’s Dictionary of Fungi. 10th edn. Wallingford: CAB International. https://doi.org/10.1079/9780851998268.0000
- Kunze, G. (1823) Einige neue oder verkannten Pilzgattungen und Arten. X. Myxotrichum. Mykologische Hefte 2: 108–110.
- Lee, J.H., Ten, L.N., Lim, S.K., Ryu, J.J., Avalos-Ruiz, D., Lee, S.Y. & Jung, H.Y. (2022) Molecular and Morphological Characteristics of a new Species Collected from an Insect (Cicindela transbaicalica) in Korea. Mycobiology 50: 181–187. https://doi.org/10.1080/12298093.2022.2080333
- Liang, J., Liu, B.Y., Li, Z.Y., Meng, W., Wang, Q.G. & Xu, L.J. (2019) Myxotrichum albicans, a new slowly-growing species isolated from forest litters in China. Mycoscience 60: 232–236. https://doi.org/10.1016/j.myc.2019.03.002
- Lin, L.C., Ye, Y.S. & Lin, W.R. (2019) Characteristics of root-cultivable endophytic fungi from Rhododendron ovatum Planch. Brazilian Journal of Microbiology 50: 185–93. https://doi.org/10.1007/s42770-018-0011-8
- Liu, D., Coloe, S., Baird, R. & Pedersen, J. (2000) Rapid mini-preparation of fungal DNA for PCR. Journal of Clinical Microbiology 38: 471. https://doi.org/10.1128/JCM.38.1.471-471.2000
- Lumley, T.C., Gignac, L.D. & Currah, R.S. (2001) Microfungus communities of white spruce and trembling aspen logs at different stages of decay in disturbed and undisturbed sites in the boreal mixed wood region of Alberta. Canadian Journal of Botany 79: 76–92. https://doi.org/10.1139/b00-135
- Matsushima, T. (1995) Matsushima Mycological Memoirs No. 8. Kobe, Japan. pp. 1–44.
- Mc Neill, J., Barrie, F.R., Buck, W.R., Demoulin, V., Greuter, W., Hawksworth, D.L., Herendeen, P.S., Knapp, S., Marhold, K., Prado, J., Prud‘Homme Van Reine, W.F., Smith, G.F., Wiersema, J.H. & Turland, N.J. (2012) International code of nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Vegetabile 154: 1–240.
- Miller, M.A. & Pfeiffer,W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), November 14, 2010. New Orleans, Louisiana, pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Min, Y.J., Park, M.S., Fong, J.J., Quan, Y., Jung, S. & Lim, Y.W. (2014) Diversity and saline resistance of endophytic fungi associated with Pinus thunbergii in coastal shelterbelts of Korea. Journal of Microbiology and Biotechnology 243: 324–333. https://doi.org/10.4014/jmb.1310.10041
- Morrall, R.A.A. (1968) Two new species of Oidiodendron from boreal forest soils. Canadian Journal of Botany 46: 203–206. https://doi.org/10.1139/b68-034
- Mucciarelli, M., Scannerini, S., Bertea, C. & Maffei, M. (2003) In vitro and in vivo peppermint (Mentha piperita) growth promotion by nonmycorrhizal fungal colonization. New Phytologist 158: 579–591. https://doi.org/10.1046/j.1469-8137.2003.00762.x
- Norvell, L.L. (2011) Fungal nomenclature. 1. Melbourne approves a new Code. Mycotaxon 116: 481–490. https://doi.org/10.5248/116.481
- Orr, G.F., Kuehn, H.H. & Plunkett, O.A. (1963) The genus Myxotrichum Kunze. Canadian Journal of Botany 41: 1457–1480. https://doi.org/10.1139/b63-127
- Parbery, D.G. (1969) Amorphotheca resinae gen. nov., sp. nov.: The perfect state of Cladosporium resinae. Australian Journal of Botany 17: 331–357. https://doi.org/10.1071/BT9690331
- Pivkin, M.V. (2000) Filamentous fungi associated with Holothurians from the Sea of Japan, off the Primorye Coast of Russia. Biological Bulletin 198: 101–109. https://doi.org/10.2307/1542808
- Rambaut, A. (2012) FigTree version 1.4.0. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 15 May 2024)
- Rangel-Grimaldo, M., Macías-Rubalcava, M.L., González-Andrade, M., Raja, H., Figueroa, M. & Mata, R. (2020) α-Glucosidase and protein tyrosine phosphatase 1B inhibitors from Malbranchea circinata. Journal of natural products 83: 675–683. https://doi.org/10.1021/acs.jnatprod.9b01108
- Rannala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10.1007/BF02338839
- Raviraja, N.S. (2005) Fungal endophytes in five medicinal plant species from Kudremukh Range, Western Ghats of India. Journal of Basic Microbiology 45: 230–235. https://doi.org/10.1002/jobm.200410514
- Rehner, S.A. & Samuels, G.J. (1995) Molecular systematics of the Hypocreales: a teleomorph gene phylogeny and the status of their anamorphs. Canadian Journal of Botany 73: S816–S823. https://doi.org/10.1139/b95-327
- Rice, A.V. & Currah, R.S. (2005) Oidiodendron: a survey of the named species and related anamorphs of Myxotrichum. Studies in Mycology 53: 83–120. https://doi.org/10.3114/sim.53.1.83
- Robak, H. (1932) Investigations regarding fungi on Norwegian ground wood pulp and fungal infections at wood pulp mills. Saertrykk av Nyt Magazin for Naturvidenskaberne 71: 185–330.
- Rodriguez, R.J., Henson, J., Van Volkenburgh, E., Hoy, M., Wright, L., Beckwith, F., Kim, Y.-O. & Redman, R.S. (2008) Stress tolerance in plants via habitat-adapted symbiosis. The ISME Journal 2: 404–416. https://doi.org/10.1038/ismej.2007.106
- Rodríguez-Andrade, E., Stchigel, A.M., Terrab, A., Guarro, J. & Cano-Lira, J.F. (2019) Diversity of xerotolerant and xerophilic fungi in honey. IMA Fungus 10: 1–30. https://doi.org/10.1186/s43008-019-0021-7
- Rodríguez-Andrade, E., Cano-Lira, J.F., Wiederhold, N., Pérez-Cantero, A., Guarro, J. & Stchigel, A.M. (2021) A revision of malbranchea-like fungi from clinical specimens in the United States of America reveals unexpected novelty. IMA fungus 12 (1): 25. https://doi.org/10.1186/s43008-021-00075-x
- Seifert, K.A., Hughes, S.J., Boulay, H. & Louis-Seize, G. (2007) Taxonomy, nomenclature and phylogeny of three cladosporium-like hyphomycetes, Sorocybe resinae, Seifertia azaleae and the Hormoconis anamorph of Amorphotheca resinae. Studies in Mycology 58: 235–245. https://doi.org/10.3114/sim.2007.58.09
- Sigler, L. & Carmichael, J.W. (1976) Taxonomy of Malbranchea and some other Hyphomycetes with arthroconidia. Mycotaxon 4 (2): 349–488.
- Smith, G. (1946) Note on the occurrence of species of Oidiodendron Robak in Britain. British Mycological Society Transactions 29: 232–234. https://doi.org/10.1016/S0007-1536(46)80005-6
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Stoyke, G. & Currah, R.S. (1991) Endophytic fungi from the mycorrhizae of alpine ericoid plants. Canadian Journal of Botany 69: 347–352. https://doi.org/10.1139/b91-047
- Sugiyama, M. & Mikawa, T. (2001) Phylogenetic analysis of the nonpathogenic genus Spiromastix (Onygenaceae) and related onygenalean taxa based on large subunit ribosomal DNA sequences. Mycoscience 42: 413–421. https://doi.org/10.1007/BF02464337
- Sugiyama, M., Ohara, A. & Mikawa, T. (1999) Molecular phylogeny of onygenalean fungi based on small subunit ribosomal DNA (SSU rDNA) sequences. Mycoscience 40: 251–258. https://doi.org/10.1007/BF02463962
- Szűcs, Z., Plaszkó, T., Cziáky, Z., Kiss-Szikszai, A., Emri, T., Bertóti, R., Sinka, L.T., Vasas, G. & Gonda, S. (2018) Endophytic fungi from the roots of horseradish (Armoracia rusticana) and their interactions with the defensive metabolites of the glucosinolate-myrosinase-isothiocyanate system. BMC Plant Biology 18: 85. https://doi.org/10.1186/s12870-018-1295-4
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biololgy and Evolution 30: 2725–2729. https://doi.org/10.1093/molbev/mst197
- Tsuneda, A. & Currah, R.S. (2004) Ascomatal morphogenesis in Myxotrichum arcticum supports the derivation of the Myxotrichaceae from a discomycetous ancestor. Mycologia 96: 627–635. https://doi.org/10.1080/15572536.2005.11832958
- Udagawa, S.-I. & Uchiyama, S. (1999) Taxonomic studies on new or critical fungi of non-pathogenic Onygenales 2. Mycoscience 40: 291–305. https://doi.org/10.1007/BF02463966
- Udagawa, S.-I. & Uchiyama, S. (2000) Taxonomic studies on new or critical fungi of non-pathogenic Onygenales 3. Mycoscience 41: 303–311. https://doi.org/10.1007/BF02463943
- Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Groenewald, J.Z., Cardinali, G., Houbraken, J., Boekhout, T., Crous, P.W., Robert, V. & Verkley, G.J.M. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. https://doi.org/10.1016/j.simyco.2018.05.001
- Wei, X., Chen, J., Zhang, C. & Pan, D. (2016) A New Oidiodendron maius Strain Isolated from Rhododendron fortunei and its Effects on Nitrogen Uptake and Plant Growth. Frontiers in Microbiology 7: 1327. https://doi.org/10.3389/fmicb.2016.01327
- White, T.J., Bruns, T., Lee, S.J. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, New York, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sanchez-Garcia, M., Goto, B.T., Saxena, R.K., Erdogdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., Błaszkowski, J., Boonyuen, N., da Silva, G.A., de Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Mešić, A., Monteiro, J.S., Oehl, F., Pawłowska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro., Tang, L.Z., Stadler, M., Yurkov, A. & Thines, M. (2022) Outline of Fungi and fungus-like taxa—2021. Mycosphere 13: 53–453. https://doi.org/10.5943/mycosphere/13/1/2
- Wynns, A.A. (2015) Convergent evolution of highly reduced fruiting bodies in Pezizomycotina suggests key adaptations to the bee habitat. BMC Evolutionary Biology 15: 145–156. https://doi.org/10.1186/s12862-015-0401-6
- Yuan, C., Wang, H.Y., Wu, C.S., Jiao, Y., Li, M., Wang, Y.Y., Wang, S.Q., Zhao, Z.T. & Lou, H.X. (2013) Austdiol, fulvic acid and citromycetin derivatives from an endolichenic fungus, Myxotrichum sp.. Phytochemistry Letters 6: 662–666. https://doi.org/10.1016/j.phytol.2013.08.011