Abstract
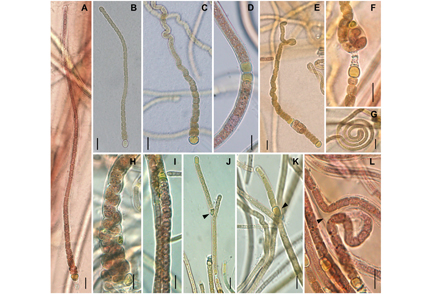
In this study, novel Calothrix-like strains NUACC09 and NUACC10 were isolated from the surfaces of rocks in Phayao Lake, Thailand. Morphological, molecular and ecological comparisons were investigated to characterize the taxonomic status of these novel strains. Under the light microscope, morphological studies indicated that these two strains were morphologically similar to Calothrix but could be differentiated by production of a non-hyaline hair at the terminal end, a low degree of tapering, twisted or loop-forming filaments, a knotted growth form, intertwined trichomes and presenting more than one trichome within the single sheath. In the 16S rRNA and rbcLX gene phylogenetic tree analyses, our strains formed a monophyletic clade with former freshwater/terrestrial Calothrix-like taxa separating distantly from other Calothrix-like genera. Furthermore, low 16S rRNA gene sequence similarity (<94.9%) to the closely related genera and species delimitation analyses (PTP/bPTP, GMYC, ABGD and ASAP methods) indicated that this clade should be considered as a different cyanobacterial lineage. The phylogenetic tree and secondary structures (D1–D1′, V2, Box-B and V3 helix) based on 16S–23S rRNA ITS regions suggested that multiple species might be contained in this clade. Therefore, here, Phayaothrix was proposed as a novel cyanobacterial genus with Phayaothrix lacustris sp. nov. as the type species following the International Code of Nomenclature for algae, fungi and plants.
References
- Abdullin, S.R., Nikulin, A.Y., Bagmet, V.B., Nikulin, V.Y. & Gontcharov, A.A. (2021) New cyanobacterium Aliterella vladivostokensis sp. nov. (Aliterellaceae, Chroococcidiopsidales), isolated from temperate monsoon climate zone (Vladivostok, Russia). Phytotaxa 527: 221–233. https://doi.org/10.11646/phytotaxa.527.3.7
- Alvarenga, D.O., Andreote, A.P.D., Branco, L.H.Z., Delbaje, E., Cruz, R.B., de Mello Varani, A. & Fiore, M.F. (2021) Amazonocrinis nigriterrae gen. nov., sp. nov., Atlanticothrix silvestris gen. nov., sp. nov. and Denronalium phyllosphericum gen. nov., nostocacean cyanobacteria from Brazilian environments. International Journal of Systematic and Evolutionary Microbiology 71: 1–12. https://doi.org/10.1099/ijsem.0.004811
- Alvarenga, D.O., Rigonato, J., Branco, L.H.Z., Melo, I.S. & Fiore, M.F. (2016) Phyllonema aviceniicola gen. nov., sp nov and Foliisarcina bertiogensis gen. nov., sp nov., epiphyllic cyanobacteria associated with Avicennia schaueriana leaves. International Journal of Systematic and Evolutionary Microbiology 66: 689–700. https://doi.org/10.1099/ijsem.0.000774
- Babić, O., Kovač, D., Rašeta, M., Šibul, F., Svirčev, Z. & Simeunović, J. (2016) Evaluation of antioxidant activity and phenolic profile of filamentous terrestrial cyanobacterial strains isolated from forest ecosystem. Journal of Applied Phycology 28: 2333–2342. https://doi.org/10.1007/s10811-015-0773-4
- Baldarelli, L.M., Pietrasiak, N., Osorio‐Santos, K. & Johansen, J.R. (2022) Mojavia aguilerae and M. dolomitestris—two new nostocaceae (cyanobacteria) species from the Americas. Journal of Phycology 58: 502–516. https://doi.org/10.1111/jpy.13275
- Berrendero, E., Perona, E. & Mateo, P. (2008) Genetic and morphological characterization of Rivularia and Calothrix (Nostocales, Cyanobacteria) from running water. International Journal of Systematic and Evolutionary Microbiology 58: 447–460. https://doi.org/10.1099/ijs.0.65273-0
- Berrendero, E., Perona, E. & Mateo, P. (2011) Phenotypic variability and phylogenetic relationships of the genera Tolypothrix and Calothrix (Nostocales, Cyanobacteria) from running water. International Journal of Systematic and Evolutionary Microbiology 61: 3039–3051. https://doi.org/10.1099/ijs.0.027581-0
- Berrendero‐Gómez, E., Johansen, J.R., Kaštovský, J., Bohunická, M. & Čapková, K. (2016) Macrochaete gen. nov.(Nostocales, Cyanobacteria), a taxon morphologically and molecularly distinct from Calothrix. Journal of Phycology 52: 638–655. https://doi.org/10.1111/jpy.12425
- Berthold, D.E., Werner, V.R., Lefler, F.W., Simon, I.P. & Laughinghouse IV, H.D. (2022) The novel marine cyanobacterium Nunduva sanctimaloensis sp. nov.(Nostocales, Cyanobacteria) from rocky shores and its reproduction through modified monocytes. Fottea 22: 192–203. https://doi.org/10.5507/fot.2021.023
- Bohunická, M., Pietrasiak, N., Johansen, J.R., Gómez, E.B., Hauer, T., Gaysina, L.A. & Lukešová, A. (2015) Roholtiella, gen. nov.(Nostocales, Cyanobacteria)—a tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa 197: 84–103. https://doi.org/10.11646/phytotaxa.197.2.2
- Bornet, E. & Flahault, C. (1886 ‘1887’) Revision des Nostocacées hétérocystées contenues dans les principaux herbiers de France (deuxième fragment). Annales des Sciences Naturelles, Botanique, Septième Série 3: 323–381; 4: 343–373 (1887).
- Brown, A.O., Romanis, C.S., Dvořák, P., Foss, A.J., Gibson, Q.A., Villanueva, C.D., Durden, W.N., Garvey, A.D., Jenkins, P., Hašler, P., Johansen, J.R., Neilan, B.A. & Casamatta, D.A. (2021) A new species of cryptic cyanobacteria isolated from the epidermis of a bottlenose dolphin and as a bioaerosol. Phycologia 60: 603–618. https://doi.org/10.1080/00318884.2021.1968173
- Cai, F.F. & Li, R.H. (2019) Validation of Compactonostoc shennongjiaense gen. et sp. nov. (Nostocaceae, Cyanobacteria). Notulae Algarum 121: 1.
- Casamatta, D.A., Villanueva, C.D., Garvey, A.D., Stocks, H.S., Vaccarino, M., Dvořák, P., Hašler, P. & Johansen, J.R. (2020) Reptodigitus chapmanii (Nostocales, hapalosiphonaceae) gen. nov.: a unique nostocalean (Cyanobacteria) genus based on a polyphasic approach. Journal of Phycology 56: 425–436. https://doi.org/10.1111/jpy.12954
- Chan, P.P., Lin, B.Y., Mak, A.J. & Lowe, T.M. (2021): tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Research 49: 9077–9096. https://doi.org/10.1093/nar/gkab688
- Choi, H.J., Joo, J.H., Kim, J.H., Wang, P., Ki, J.S. & Han, M.S. (2018) Morphological characterization and molecular phylogenetic analysis of Dolichospermum hangangense (Nostocales, Cyanobacteria) sp. nov. from Han River, Korea. Algae 33: 143–156. https://doi.org/10.4490/algae.2018.33.5.2
- Cordeiro, R., Luz, R., Vasconcelos, V., Gonçalves, V. & Fonseca, A. (2020) Cyanobacteria phylogenetic studies reveal evidence for polyphyletic genera from thermal and freshwater habitats. Diversity 12: 298. https://doi.org/10.3390/d12080298
- Domínguez-Escobar, J., Beltrán, Y., Bergman, B., Díez, B., Ininbergs, K., Souza, V. & Falcón, L.I. (2011) Phylogenetic and molecular clock inferences of cyanobacterial strains within Rivulariaceae from distant environments. FEMS Microbiology Letters 316: 90–99. https://doi.org/10.1111/j.1574-6968.2010.02195.x
- Dvořák, P., Hašler, P., Casamatta, D.A. & Poulíčková, A. (2021) Underestimated cyanobacterial diversity: trends and perspectives of research in tropical environments. Fottea 21: 110–127. https://doi.org/10.5507/fot.2021.009
- Dvořák, P., Poulíčková, A., Hašler, P., Belli, M., Casamatta, D.A. & Papini, A. (2015) Species concepts and speciation factors in cyanobacteria, with connection to the problems of diversity and classification. Biodiversity and Conservation 24: 739–757. https://doi.org/10.1007/s10531-015-0888-6
- Dvořák, P., Jahodářová, E., Casamatta, D.A., Hašler, P. & Poulíčková, A. (2018) Difference without distinction? Gaps in cyanobacterial systematics, when more is just too much. Fottea 18: 130–136. https://doi.org/10.5507/fot.2017.023
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 32: 1792–1797. https://doi.org/10.1093/nar/gkh340
- Erwin, P.M. & Thacker, R.W. (2008) Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Molecular Ecology 17: 2937–2947. https://doi.org/10.1111/j.1365-294X.2008.03808.x
- Foster, R.A., Subramaniam, A. & Zehr, J.P. (2009) Distribution and activity of diazotrophs in the Eastern Equatorial Atlantic. Environmental Microbiology 11: 741–750. https://doi.org/10.1111/j.1462-2920.2008.01796.x
- Fujisawa, T. & Barraclough, T.G. (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Systematic Biology 62: 707–724. https://doi.org/10.1093/sysbio/syt033
- Gkelis, S., Panou, M., Konstantinou, D., Apostolidis, P., Kasampali, A., Papadimitriou, S., Kati, D., Lorenzo, G.M.D., Loakeim, S., Zervou, S-K., Christophoridis, C., Triantis, T.M., Kaloudis, T., Hiskia, A. & Arsenakis, M. (2019) Diversity, cyanotoxin production, and bioactivities of cyanobacteria isolated from freshwaters of Greece. Toxins 11: 436. https://doi.org/10.3390/toxins11080436
- González-Resendiz, L., Johansen, J.R., Alba-Lois, L., Segal-Kischinevzky, C., Escobar-Sanchez, V., Jimenez-Garcia, L.F., Hauer, T. & León-Tejara, H. (2018) Nunduva, a new marine genus of Rivulariaceae (Nostocales, Cyanobacteria) from marine rocky shores. Fottea 18: 86–105. https://doi.org/10.5507/fot.2017.018
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. https://doi.org/10.1093/sysbio/syq010
- Hauer, T., Bohunická, M. & Muehlsteinova, R. (2013) Calochaete gen. nov. (Cyanobacteria, Nostocales), a new cyanobacterial type from the “páramo” zone in Costa Rica. Phytotaxa 109: 36–44. https://doi.org/10.11646/phytotaxa.109.1.4
- Hauer, T., Bohunická, M., Johansen, J.R., Mareš, J. & Berrendero‐Gomez, E. (2014) Reassessment of the cyanobacterial family Microchaetaceae and establishment of new families Tolypothrichaceae and Godleyaceae. Journal of Phycology 50: 1089–1100. https://doi.org/10.1111/jpy.12241
- Hentschke, G.S., Johansen, J.R., Pietrasiak, N., Fiore, M.D.F., Rigonato, J., Sant’Anna, C.L. & Komarek, J. (2016) Phylogenetic placement of Dapisostemon gen. nov. and Streptostemon, two tropical heterocytous genera (Cyanobacteria). Phytotaxa 245: 129–143. https://doi.org/10.11646/phytotaxa.245.2.4
- Iteman, I., Rippka, R., Tandeau de Marcac, N. & Herdmann, M. (2000) Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146: 1275–1286. https://doi.org/10.1099/00221287-146-6-1275
- Johansen, J.R. & Casamatta, D.A. (2005) Recognizing cyanobacterial diversity through adoption of a new species paradigm. Algological Studies 117: 71–93. https://doi.org/10.1127/1864-1318/2005/0117-0071
- Johansen, J.R., González‐Resendiz, L., Escobar‐Sánchez, V., Segal‐Kischinevzky, C., Martínez‐Yerena, J., Hernández‐Sánchez, J., Hernández-Pérez, G. & León‐Tejera, H. (2021) When will taxonomic saturation be achieved? A case study in Nunduva and Kyrtuthrix (Rivulariaceae, Cyanobacteria). Journal of Phycology 57: 1699–1720. https://doi.org/10.1111/jpy.13201
- Johansen, J.R., Mareš, J., Pietrasiak, N., Bohunická, M., Zima Jr, J., Štenclová, L. & Hauer, T. (2017) Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria). PLoS One 12: e0186393. https://doi.org/10.1371/journal.pone.0186393
- Kaštovský, J., Berrendero-Gomez, E., Hladil, J. & Johansen, J.R. (2014) Cyanocohniella calida 673 gen. nov. et spec. nov. (Cyanobacteria: Aphanizomenonaceae) a new cyanobacterium from the thermal springs from Karlovy Vary, Czech Republic. Phytotaxa 181: 279–292. https://doi.org/10.11646/phytotaxa.181.5.3
- Komárek, J. (2016) A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. European Journal of Phycology 51: 346–353. https://doi.org/10.1080/09670262.2016.1163738
- Komárek, J. (2023) Taxonomic review of cyanobacteria 2021/2022 according to polyphasic evaluation. Fottea 23: 141–148. https://doi.org/10.5507/fot.2022.017
- Komárek, J., Kaštovský, J., Mareš, J. & Johansen, J.R. (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86: 295–335. [https://www.preslia.cz/article/103]
- Kumar, M., Prasanna, R., Bidyarani, N., Babu, S., Mishra, B.K., Kumar, A., Adak, A., Jauhari, S., Yadav, K., Singh, R. & Saxena, A.K. (2013) Evaluating the plant growth promoting ability of thermotolerant bacteria and cyanobacteria and their interactions with seed spice crops. Scientia Horticulturae 164: 94–101. https://doi.org/10.1016/j.scienta.2013.09.014
- Kumar, N., Saraf, A., Pal, S., Mishra, D., Singh, P. & Johansen, J.R. (2023) Circumscription of Fulbrightiella gen. nov. and Sherwoodiella gen. nov., Two Novel Genera in the Calotrichaceae (Nostocales, Cyanobacteria). Journal of Phycology 59: 204–220. https://doi.org/10.1016/j.scienta.2013.09.014
- Mai, T., Johansen, J.R., Pietrasiak, N., Bohunická, M. & Martin, M.P. (2018) Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 365: 1–59. https://doi.org/10.11646/phytotaxa.365.1.1
- Mareš, J., Johansen, J.R., Hauer, T., Zima Jr, J., Ventura, S., Cuzman, O., Tiribilli, B. & Kaštovský, J. (2019) Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea, and Zehria. Journal of Phycology 55: 578–610. https://doi.org/10.1111/jpy.12853
- Mathews, D.H. (2014) RNA secondary structure analysis using RNAstructure. Current Protocols in Bioinformatics 46: 1–25. https://doi.org/10.1002/0471250953.bi1206s46
- Mishra, D., Saraf, A., Kumar, N., Pal, S. & Singh, P. (2021) Issues in cyanobacterial taxonomy: comprehensive case study of unbranched, false branched and true branched heterocytous cyanobacteria. FEMS Microbiology Letters 368: fnab005. https://doi.org/10.1093/femsle/fnab005
- Nowruzi, B. & Soares, F. (2021) Alborzia kermanshahica gen. nov., sp. nov. (Chroococcales, Cyanobacteria), isolated from paddy fields in Iran. International Journal of Systematic and Evolutionary Microbiology 71: 004828. https://doi.org/10.1099/ijsem.0.004828
- Nylander, J.A.A. (2004) MrModeltest, vol 2. Uppsala University, Program distributed by the author. Evolutionary Biology Centre
- Osorio-Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Kováčik, L., Martin, M.P. & Johansen, J.R. (2014) Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. European journal of phycology 49: 450–470. https://doi.org/10.1080/09670262.2014.976843
- Okonechnikov, K., Golosova, O., Fursov, M. & Ugene Team. (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167. https://doi.org/10.1093/bioinformatics/bts091
- Pathak, J., Maurya, P.K., Singh, S.P., Häder, D.P. & Sinha, R.P. (2018) Cyanobacterial farming for environment friendly sustainable agriculture practices: innovations and perspectives. Frontiers in Environmental Science 6: 7. https://doi.org/10.3389/fenvs.2018.00007
- Pietrasiak, N., Osorio‐Santos, K., Shalygin, S., Martin, M.P. & Johansen, J.R. (2019) When is a lineage a species? A case study in Myxacorys gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. Journal of Phycology 55: 976–996. https://doi.org/10.1111/jpy.12897
- Pietrasiak, N., Reeve, S., Osorio‐Santos, K., Lipson, D.A. & Johansen, J.R. (2021) Trichotorquatus gen. nov.‐a new genus of soil cyanobacteria discovered from American drylands1. Journal of Phycology 57: 886–902. https://doi.org/10.1111/jpy.13147
- Puillandre, N., Brouillet, S. & Achaz, G. (2021): ASAP assemble species by automatic partitioning. Molecular Ecology Resources 21: 609–620. https://doi.org/10.1111/1755-0998.13281
- Puillandre, N., Lambert, A., Brouillet, S. & Achaz, G.J.M.E. (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21: 1864–1877. https://doi.org/10.1111/j.1365-294x.2011.05239.x
- R Development Core Team (2009) R: A language and environment for statistical computing. Vienna, Austria. (ISBN 3-900051-07-0).
- Rasouli-Dogaheh, S., Noroozi, M., Khansha, J., Amoozegar, M.A., Rahimi, R., Nikou, M.M. & Hauer, T. (2023) Khargia gen. nov., a new genus of simple trichal Cyanobacteria from the Persian Gulf. Fottea 23: 49–61. https://doi.org/10.5507/fot.2022.012
- Řeháková, K., Johansen, J.R., Casamatta, D.A., Xuesong, L. & Vincent, J. (2007) Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. Phycologia 46: 481–502. https://doi.org/10.2216/06-92.1
- Renuka, N., Sood, A., Ratha, S.K., Prasanna, R. & Ahluwalia, A.S. (2013) Nutrient sequestration, biomass production by microalgae and phytoremediation of sewage water. International Journal of Phytoremediation 15: 789–800. https://doi.org/10.1080/15226514.2012.736436
- Rippka, R., Waterbury, J. & Cohen-Bazire, G. (1974) A cyanobacterium which lacks thylakoids. Archives for Microbiology 100: 419–436. https://doi.org/10.1007/BF00446333
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Saraf, A.G., Dawda, H.G. & Singh, P. (2019a) Validation of the genus Desikacharya gen. nov. (Nostocaceae, Cyanobacteria) and three included species. Notulae Algarum 107: 1–3.
- Saraf, A., Suradkar, A., Dawda, H.G., Gaysina, L.A., Gabidullin, Y., Kumat, A., Behere, I., Kotulkar, M., Batule, P. & Singh, P. (2019b) Phylogenetic complexities of the members of Rivulariaceae with the re-creation of the family Calotrichaceae and description of Dulcicalothrix necridiiformans gen nov., sp nov., and reclassification of Calothrix desertica. FEMS Microbiology Letters 366: fnz219. https://doi.org/10.1093/femsle/fnz219
- Sihvonen, L.M., Lyra, C., Fewer, D.P., Rajaniemi-Wacklin, P., Lehtimäki, J.M., Wahlsten, M. & Sivonen, K. (2007) Strains of the cyanobacterial genera Calothrix and Rivularia isolated from the Baltic Sea display cryptic diversity and are distantly related to Gloeotrichia and Tolypothrix. FEMS Microbiology Ecology 61: 74–84. https://doi.org/10.1111/j.1574-6941.2007.00321.x
- Shalygin, S., Shalygina, R., Johansen, J.R., Pietrasiak, N., Berrendero, G.E., Bohunická, M., Mareš, J. & Sheil, C.A. (2017) Cyanomargarita gen. nov.(Nostocales, Cyanobacteria): convergent evolution resulting in a cryptic genus. Journal of Phycology 53: 762–777. https://doi.org/10.1111/jpy.12542
- Smith, C., Heyne, S., Richter, A.S., Will, S. & Backofen, R. (2010) Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LocARNA. Nucleic Acids Research 38: 7W373–W377. https://doi.org/10.1093/nar/gkq316
- Stackebrandt, E. & Ebers, J. (2006) Taxonomic Parameters Revisited: Tarnished Gold Standards. Microbiology Today 33: 152–155. [Available on line https://microbiologysociety.org/publication/past-issues/systematics.html]
- Stackebrandt, E. & Goebel, B.M. (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology 44: 846–849. https://doi.org/10.1099/00207713-44-4-846
- Strunecký, O., Ivanova, A.P. & Mareš, J. (2023) An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. Journal of Phycology 59: 12–51. https://doi.org/10.1111/jpy.13304
- Tawong, W., Pongcharoen, P. & Saijuntha, W. (2022a) Neocylindrospermum variakineticum gen. & sp. nov. (Nostocales, Cyanobacteria), a novel genus separated from Cylindrospermum using a polyphasic method. Phycologia 61: 653–668. https://doi.org/10.1080/00318884.2022.2130829
- Tawong, W., Pongcharoen, P., Nishimura, T. & Saijuntha, W. (2022b) Siamcapillus rubidus gen. et sp. nov.(Oculatellaceae), a novel filamentous cyanobacterium from Thailand based on molecular and morphological analyses. Phytotaxa 558: 33–52. https://doi.org/10.11646/phytotaxa.558.1.2
- Tuo, S.H., Mulholland, M.R., Taniuchi, Y., Chen, H.Y., Jane, W.N., Lin, Y.H. & Chen, Y.L.L. (2021) Trichome lengths of the heterocystous N2-Fixing cyanobacteria in the tropical marginal seas of the Western North Pacific. Frontiers in Marine Science 8: 678607. https://doi.org/10.3389/fmars.2021.678607
- Uher, B. (2007): Morphological characterization of three subaerial Calothrix species (Nostocales, Cyanobacteria). Fottea 1: 33–38. https://doi.org/10.5507/fot.2007.002
- Vaccarino, M.A. & Johansen, J.R. (2012) Brasilonema Angustatum sp. nov. (Nostocales), a new filamentous cyanobacterial species from the Hawaiian Islands. Journal of Phycology 48: 1178–1186. https://doi.org/10.1111/j.1529-8817.2012.01203.x
- Villanueva, C.D., Garvey, A.D., Hašler, P., Dvořák, P., Poulíčková, A., Norwich, A.R. & Casamatta, D.A. (2019) Descriptions of Brasilonema geniculatum and Calothrix dumus (Nostocales, Cyanobacteria): two new taxa isolated from cemetery tombstones. Phytotaxa 387: 1–20. https://doi.org/10.11646/phytotaxa.387.1.1
- Ward, R.D., Stajich, J.E., Johansen, J.R., Huntemann, M., Clum, A., Foster, B., Foster, B., Roux, S., Palaniappan, K., Varghese, N., Mukherjee, S., Reddy, T.B.K., Daum, C., Copeland, A., Chen, I.-M.A., Ivanova, N.N., Kyrpides, N.C., Shapiro, N., Eloe-Fadrosh, E.A. & Pietrasiak, N. (2021) Metagenome sequencing to explore phylogenomics of terrestrial cyanobacteria. Microbiology Resource Announcements 10: e00258-21. https://doi.org/10.1128/mra.00258-21
- Wehr, J.D. & Sheath, R.G. (2003) Freshwater habitats of algae. In: Wehr, J.D. & Sheath, R.G. (eds.) Freshwater Algae of North America: Ecology and Classification. Academic Press, San Diego, CA, pp. 11–59. https://doi.org/10.1016/B978-012741550-5/50003-9
- West, W. & West, G.S. (1897) Welwitsch’s African freshwater algae. Journal of Botany, British and Foreign 35: 1–7.
- Wilmotte, A., Laughinghouse IV, H.D., Capelli, C., Rippka, R. & Salmaso, N. (2017) Taxonomic identification of cyanobacteria by a polyphasic approach. In: Kurmayer, R., Sivonen, K., Wilmotte, A. & Salmaso, N. (Eds.) Molecular tools for the detection and quantification of toxigenic cyanobacteria. Wiley, NJ, pp. 79–134. https://doi.org/10.1002/9781119332169.ch4
- Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.H., Whitman, W.B., Euzéby, J., Amann, R. & Rosselló-Móra, R. (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Reviews Microbiology 12: 635–645. https://doi.org/10.1038/nrmicro3330
- Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876. https://doi.org/10.1093/bioinformatics/btt499