Abstract
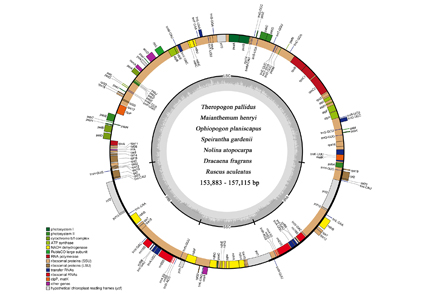
Convallarioideae are a subfamily of Asparagaceae, a morphologically diverse group comprising seven tribes, i.e., Eriospermeae, Rusceae, Dracaeneae, Nolineae, Convallarieae, Ophiopogoneae and Polygonateae, and two genera unclassified to tribe. In this study, we conducted comparative plastid genome (plastome) analyses of seven species representing seven clades of the subfamily. The results showed that all plastomes exhibited standard conserved quadripartite structures with inverted repeats (26,261–26,522 bp) separated by a large single-copy region (83,007–85,692 bp) and a small single-copy region (18,205–18,707 bp). Each plastome has 137 genes, including 87 protein-coding genes, 38 transfer RNAs and 8 ribosomal RNA genes. Furthermore, we detected 33–61 simple sequence repeats in six categories and 39–57 long repeats in four categories. We selected eleven hotspots as primer sites for potential molecular markers. Phylogenetic analysis of the subfamily revealed that all the tribes are strongly supported except for Polygonateae, but weak support was observed among the main clades in the subfamily.
References
- Ahmad, W., Asaf, S., Khan, A., Al-Harrasi, A., Al-Okaishi, A. & Khan, A.L. (2022) Complete chloroplast genome sequencing and comparative analysis of threatened dragon trees Dracaena serrulata and Dracaena cinnabari. Scientific Reports 12: 16787. https://doi.org/10.1038/s41598-022-20304-6
- Amiryousefi, A., Hyvönen, J. & Poczai, P. (2018) IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34: 3030–3031. https://doi.org/10.1093/bioinformatics/bty220
- APG III (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. APG III. Botanical Journal of the Linnean Society 161: 105–121 https://doi.org/10.1111/j.1095-8339.2009.00996.x
- APG IV (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 141: 399–436. https://doi.org/10.1111/boj.12385
- Baillon, H. (1894) Histoire des plantes, vol. 12. Hachette, Paris, 524 pp.
- Baker, G.J. (1886) Hooker’s icones plantarum, vol. 16. Longman, Rees, Orme, Brown, Green & Longman, London, 1537 pp.
- Baker, G.J. (1889) Hooker’s icones plantarum, vol. 19. Longman, Rees, Orme, Brown, Green & Longman, London, 1867 pp.
- Bartlett, H.H. (1909) Nolina in the southern Atlantic states. Rhodora 11: 80–81.
- Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33: 2583–2585. https://doi.org/10.1093/bioinformatics/btx198
- Bolger, A.M., Lohse, M. & Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. https://doi.org/10.1093/bioinformatics/btu170
- Chen, M.M., Zhang, M., Liang, Z.S. & He, Q.L. (2022) Characterization and comparative analysis of chloroplast genomes in five Uncaria species endemic to China. International Journal of Molecular Sciences 23: 11617. https://doi.org/10.3390/ijms231911617
- Chen, S., Kim, D.K., Chase, M.W. & Kim, J.H. (2013) Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLOS ONE 8: e59472. https://doi.org/10.1371/journal.pone.0059472
- Chumley, T.W., Palmer, J.D., Mower, J.P., Fourcade, H.M., Calie, P.J., Boore, J.L. & Jansen, R.K. (2006) The complete chloroplast genome sequence of Pelargonium ×hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution 23: 2175–2190. https://doi.org/10.1093/molbev/msl089
- Darling, A.C., Mau, B., Blattner, F.R. & Perna, N.T. (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Research 14: 1394–1403. https://doi.org/10.1101/gr.2289704
- Dong, W.P., Xu, C., Li, C.H., Sun, J.H., Zuo, Y.J., Shi, S., Cheng, T., Guo, J.J. & Zhou, S.L. (2015) ycf1, the most promising plastid DNA barcode of land plants. Scientific Reports 5: 8348. https://doi.org/10.1038/srep08348
- Doyle, J. (1991) DNA protocols for plants. In: Hewitt, G.M., Johnston, A.W.B. & Young, J.P.W. (Eds.) Molecular techniques in taxonomy. Springer, Berlin, pp. 283–293. https://doi.org/10.1007/978-3-642-83962-7_18
- Frazer, K.A., Pachter, L., Poliakov, A., Rubin, E.M. & Dubchak, I. (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Research 32 (suppl_2): W273–W279. https://doi.org/10.1093/nar/gkh458
- He, B., Dong, H., Jiang, C., Cao, F., Tao, S. & Xu, L.A. (2016) Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending. Scientific Reports 6: 35927. https://doi.org/10.1038/srep35927
- Hong, Z., Wu, Z., Zhao, K., Yang, Z., Zhang, N., Guo, J., Tembrock, L.R. & Xu, D. (2020) Comparative analyses of five complete chloroplast genomes from the genus Pterocarpus (Fabacaeae). International Journal of Molecular Sciences 21: 3758. https://doi.org/10.3390/ijms21113758
- Huotari, T. & Korpelainen, H. (2012) Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene 508: 96–105. https://doi.org/10.1016/j.gene.2012.07.020
- Jang, C.G. & Pfosser, M. (2002) Phylogenetics of Ruscaceae sensu lato based on rbcL and trnL-F DNA sequence data. Stapfia 85: 333–348 https://doi.org/10.1016/s0076-6879(05)95020-9
- Ji, Y.H., Landis, J.B., Yang, J., Wang, S.Y., Zhou, N., Luo, Y. & Liu, H.Y. (2023) Phylogeny and evolution of Asparagaceae subfamily Nolinoideae: new insights from plastid phylogenomics. Annals of Botany 131: 301–312. https://doi.org/10.1093/aob/mcac144
- Jin, J.J., Yu, W.B., Yang, J.B., Song, Y., Depamphilis, C.W., Yi, T.S. & Li, D.Z. (2020) GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology 21: 1–31. https://doi.org/10.1186/s13059-020-02154-5
- Kalia, R.K., Rai, M.K., Kalia, S., Singh, R. & Dhawan, A.K. (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177: 309–334. https://doi.org/10.1007/s10681-010-0286-9
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010
- Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P. & Drummond, A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
- Ker Gawler & Bellende, J. (1808) Dracaena fragrans. Botanical Magazine 27: pl. 1081.
- Khakhlova, O. & Bock, R. (2006) Elimination of deleterious mutations in plastid genomes by gene conversion. The Plant Journal 46: 85–94. https://doi.org/10.1111/j.1365-313x.2006.02673.x
- Kim, J.H., Kim, D.K., Forest, F., Fay, M.F. & Chase, M.W. (2010) Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Annals of Botany 106: 775–790. https://doi.org/10.1093/aob/mcq167
- Kurtz, S., Choudhuri, J.V., Ohlebusch, E., Schleiermacher, C., Stoye, J. & Giegerich, R. (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research 29: 4633–4642. https://doi.org/10.1093/nar/29.22.4633
- Lafrankie, J.V. (1986) Transfer of the species of Smilacina to Maianthemum (Liliaceae). Taxon 35: 584–589. https://doi.org/10.2307/1221922
- Li, D.M., Pan, Y.G., Liu, H.L., Yu, B., Huang, D. & Zhu, G.F. (2024) Thirteen complete chloroplast genomes of the Costaceae family: insights into genome structure, selective pressure and phylogenetic relationships. BMC Genomics 25: 68. https://doi.org/10.1186/s12864-024-09996-4
- Li, Y.T., Dong, Y., Liu, Y.C., Yu, X.Y., Yang, M.S. & Huang, Y.R. (2021) Comparative analyses of Euonymus chloroplast genomes: genetic structure, screening for loci with suitable polymorphism, positive selection genes, and phylogenetic relationships within Celastrineae. Frontiers in Plant Science 11: 593984. https://doi.org/10.3389/fpls.2020.593984
- Li, Z., Long, H., Zhang, L., Liu, Z., Cao, H., Shi, M. & Tan, X. (2017) The complete chloroplast genome sequence of tung tree (Vernicia fordii): organization and phylogenetic relationships with other angiosperms. Scientific Reports 7: 1869. https://doi.org/10.1038/s41598-017-02076-6
- Linnaeus, C. (1753) Species plantarum, vol. 2. Salvius, Stockholm, 1041 pp.
- Linnaeus, C. (1762) Species plantarum, ed. 2, vol. 1. Salvius, Stockholm, 456 pp.
- Lohse, M., Drechsel, O. & Bock, R. (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics 52: 267–274. https://doi.org/10.1007/s00294-007-0161-y
- Long, L.X., Li, Y.T., Wang, S.J., Liu, Z.L., Wang, J.M. & Yang, M.S. (2023) Complete chloroplast genomes and comparative analysis of Ligustrum species. Scientific Reports 13: 212. https://doi.org/10.1038/s41598-022-26884-7
- Loureiro, J.D. (1790) Flora Cochinchinensis, vol. 1. Academic, Lisbon, 216 pp.
- Lu, Q.X., Chang, X., Gao, J., Wu, X., Wu, J., Qi, Z.C., Wang, R.H., Yan, X.L. & Li, P. (2022) Evolutionary comparison of the complete chloroplast genomes in Convallaria species and phylogenetic study of Asparagaceae. Genes 13: 1724. https://doi.org/10.3390/genes13101724
- Maximowicz, J.C. (1871) Les espèces d’ophioponis dans les herbiers de St- Pétersbourg. Bulletin de l’Academie Imperiale des Sciences de St-Petersbourg 10: 8390
- Meng, R., Luo, L.Y., Zhang, J.Y., Zhang, D.G., Nie, Z.L. & Meng, Y. (2021) The deep evolutionary relationships of the morphologically heterogeneous Nolinoideae (Asparagaceae) revealed by transcriptome data. Frontiers in Plant Science 11: 584981. https://doi.org/10.3389/fpls.2020.584981
- Meng, Y., Nie, Z.L., Deng, T., Wen, J. & Yang, Y.P. (2014) Phylogenetics and evolution of phyllotaxy in the Solomon’s seal genus Polygonatum (Asparagaceae: Polygonateae). Botanical Journal of the Linnean Society 176: 435–451. https://doi.org/10.1111/boj.12218
- Merrill, E.D. (1908) Flore générale de l’Indo-Chine, vol. 6. Masson, Paris, 780 pp.
- Nakai, T. (1920) Catalogus seminum et sporarum in Horto Botanico Universitatis Imperialis Tokyoensis per annos 1915 et 1916 lectorum. Tōkyō Daigaku Botanic Garden, Tokyo, 33 pp.
- Park, I., Song, J.H., Yang, S., Chae, S. & Moon, B. (2021) Plastid phylogenomic data offers novel insights into the taxonomic status of the Trichosanthes kirilowii complex (Cucurbitaceae) in south Korea. Frontiers in Plant Science 12: 559511. https://doi.org/10.3389/fpls.2021.559511
- Parvathy, S.T., Udayasuriyan, V. & Bhadana, V. (2022) Codon usage bias. Molecular Biology Reports 49: 539–565. https://doi.org/10.1007/s11033-021-06749-4
- Posada, D. & Crandall, K.A. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817
- Raman, G., Lee, E.M. & Park, S. (2021) Intracellular DNA transfer events restricted to the genus Convallaria within the Asparagaceae family: possible mechanisms and potential as genetic markers for biographical studies. Genomics 113: 2906–2918. https://doi.org/10.1016/j.ygeno.2021.06.033
- Raman, G. & Park, S. (2020) The complete chloroplast genome sequence of the Speirantha gardenii: comparative and adaptive evolutionary analysis. Agronomy 10: 1405. https://doi.org/10.3390/agronomy10091405
- Raubeson, L.A., Peery, R., Chumley, T.W., Dziubek, C., Fourcade, H.M., Boore, J.L. & Jansen, R.K. (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8: 174. https://doi.org/10.1186/1471-2164-8-174
- Rozas, J., Ferrer-Mata, A., Sánchez-Delbarrio, J.C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S.E. & Sánchez-Gracia, A. (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34: 3299–3302. https://doi.org/10.1093/molbev/msx248
- Rudall, P.J., Conran, J.G. & Chase, M.W. (2000) Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Botanical Journal of the Linnean Society 134: 73–92. https://doi.org/10.1006/bojl.2000.0365
- Seberg, O., Petersen, G., Davis, J.I., Pires, J.C., Stevenson, D.W., Chase, M.W., Fay, M.F., Devey, D.S., Jørgensen, T., Sytsma, K.J. & Pillon, Y. (2012) Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. American Journal of Botany 99: 875–889. https://doi.org/10.3732/ajb.1100468
- She, R.X., Zhao, P., Zhou, H.J., Yue, M., Yan, F., Hu, G., Gao, X.X. & Zhang, S.X. (2020) Complete chloroplast genomes of Liliaceae (s.l.) species: comparative genomic and phylogenetic analyses. Nordic Journal of Botany 38: e02477. https://doi.org/10.1111/njb.02477
- Shi, L.C., Chen, H.M., Jiang, M., Wang, L.Q., Wu, X., Huang, L.F. & Liu, C. (2019) CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Research 47: W65–W73. https://doi.org/10.1093/nar/gkz345
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Stevens, P.F. (2016) Angiosperm phylogeny website, version 13. Available from: http://www.mobot.org/MOBOT/Research/APweb/welcome.html (accessed: 26 February 2024).
- Sukhramani, G., Maurya, S. & Choudhary, R.K. (2024) Plastome comparison reveals hotspots of nucleotide diversity and positive selection pressure on accD, matK, psaA and rbcL genes in Smilacaceae. Brazilian Journal of Botany. [17 pp.] https://doi.org/10.1007/s40415-023-00973-x
- Tanaka, N. (2003) New combinations in Rohdea (Convallariaceae). Novon 13: 331. https://doi.org/10.2307/3393269
- Tanaka, N. & Nguyen, K.S. (2023) Nolinoideae (Asparagaceae) in APG III needs replacing with Convallarioideae. Phytotaxa 583: 297–299. https://doi.org/10.11646/phytotaxa.583.3.9
- Wang, J., Qian, J., Jiang, Y., Chen, X.C., Zheng, B.J., Chen, S.L., Yang, F.J., Xu, Z.C. & Duan, B.Z. (2022) Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of Polygonatum and tribe Polygonateae. Frontiers in Plant Science 13: 882189. https://doi.org/10.3389/fpls.2022.882189
- Zhu, T., Zhang, L., Chen, W., Yin, J. & Li, Q. (2017) Analysis of chloroplast genomes in 1342 plants. Genomics and Applied Biology 36: 4323–4333. https://doi.org/10.13417/j.gab.036.004323