Abstract
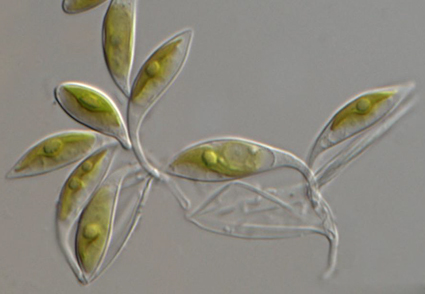
Obliquicauda belongs to the Choricystis/Botryococcus clade (Trebouxiophyceae, Chlorophyta). The establishment of this genus was predicated on phylogenetic analyses and the distinctive morphology characteristics, slightly asymmetrical and fusiform vegetative cells with a tail-like stalk tilting to one side. Two different pure cultured Obliquicauda-like strains obtained from a living Ficus pumila L. leaf were deeply investigated in this study. Based on the significant morphological differences (cell length, width, L:W ratio, and stalk length), the results of phylogenetic analyses relying on 18S and rbcL markers, and the genetic distances of the 18S and rbcL genes, these two strains were conclusively identified as novel additions to Obliquicauda, O. longipens sp. nov., and O. xianninga sp. nov.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W. & Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25 (17): 3389–3402. https://doi.org/10.1093/nar/25.17.3389
- Capella-Gutierrez, S., Silla-Martinez, J.M. & Gabaldon, T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15): 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
- Davey, M., Norman, L., Sterk, P., Huete-Ortega, M., Bunbury, F., Loh, B.K.W., Stockton, S., Peck, L.S., Convey, P., Newsham, K.K. & Smith, A.G. (2019) Snow algae communities in Antarctica: metabolic and taxonomic composition. New Phytologist 222 (3): 1242–1255. https://doi.org/10.1111/nph.15701
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 (8): 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Katoh, K. & Standley, D.M. (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30 (4): 772–780. https://doi.org/10.1093/molbev/mst010
- Kumar, V., Dey, A. & Singh, A. (2009) MEGA: A Bio Computational Software for Sequence and Phylogenetic Analysis. Paper presented at the World Congress on Engineering 2009, Imperial Coll London, London, ENGLAND.
- Li, S.Y., Sun, H., Hu, Y.X., Liu, B.W., Zhu, H., Hu, Z.Y. & Liu, G.X. (2020a) Four New Members of Foliicolous Green Algae Within the Watanabea Clade (Trebouxiophyceae, Chlorophyta) from China. Journal of Eukaryotic Microbiology 67 (3): 369–382. https://doi.org/10.1111/jeu.12787
- Li, S.Y., Tan, H.C., Liu, B.W., Zhu, H., Hu, Z.Y. & Liu, G.X. (2021) Watanabeales ord. nov. and twelve novel species of Trebouxiophyceae (Chlorophyta). Journal of Phycology 57 (4): 1167–1186. https://doi.org/10.1111/jpy.13165
- Li, S.Y., Zhu, H., Hu, Y.X., Hu, Z.Y. & Liu, G.X. (2020b) Obliquicauda gen. nov. (Trebouxiophyceae, Chlorophyta), including O. inflata sp. nov. and O. apiculata sp. nov.: foliicolous algae from Ficus leaves. Phycologia 59 (1): 35–44. https://doi.org/10.1080/00318884.2019.1667168
- Linnaeus, C. (1753) Species Plantarum 2. Imprensis Laurentii Salvii, Holmiae [Stockholm], 640 pp.
- Liu, B.W., Li, S.Y., Zhu, H. & Liu, G.X. (2023) Phyllosphere eukaryotic microalgal communities in rainforests: Drivers and diversity. Plant Diversity 45 (1): 45–53. https://doi.org/10.1016/j.pld.2022.08.0062468-2659
- Medlin, L., Elwood, H.J., Stickel, S. & Sogin, M.L. (1988) The Characterization of Enzymatically Amplified Eukaryotic 16s-Like rRNA-Coding Regions. Gene 71 (2): 491–499. https://doi.org/10.1016/0378-1119(88)90066-2
- Muggia, L., Leavitt, S. & Barreno, E. (2018) The hidden diversity of lichenised Trebouxiophyceae (Chlorophyta). Phycologia 57 (5): 503–524. https://doi.org/10.2216/17-134.1
- Neustupa, J. & Škaloud, P. (2008) Diversity of subaerial algae and cyanobacteria on tree bark in tropical mountain habitats. Biologia 63 (6): 806–812. https://doi.org/10.2478/s11756-008-0102-3
- Neustupa, J. & Škaloud, P. (2010) Diversity of subaerial algae and cyanobacteria growing on bark and wood in the lowland tropical forests of Singapore. Plant Ecology and Evolution 143 (1): 51–62. https://doi.org/10.5091/plecevo.2010.417
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution 32 (1): 268–274. https://doi.org/10.1093/molbev/msu300
- Rindi, F., Guiry, M.D. & Lopez-Bautista, J.M. (2006) New records of Trentepohliales (Ulvophyceae, Chlorophyta) from Africa. Nova Hedwigia 83 (3–4): 431–449. https://doi.org/10.1127/0029-5035/2006/0083-0431
- Rippin, M., Borchhardt, N., Williams, L., Colesie, C., Jung, P., Budel, B., Becker, B. (2018) Genus richness of microalgae and Cyanobacteria in biological soil crusts from Svalbard and Livingston Island: morphological versus molecular approaches. Polar Biology 41 (5): 909–923. https://doi.org/10.1007/s00300-018-2252-2
- Sanders, W.B., Pérez-Ortega, S., Nelsen, M.P., Lücking, R. & de los Ríos, A. (2016) Heveochlorella (Trebouxiophyceae): a little-known genus of unicellular green algae outside the Trebouxiales emerges unexpectedly as a major clade of lichen photobionts in foliicolous communities. Journal of Phycology 52 (5): 840–853. https://doi.org/10.1111/jpy.12446
- Sanders, W.B. (2001) Preliminary light microscope observations of fungal and algal colonization and lichen thallus initiation on glass slides placed near foliicolous lichen communities within a lowland tropical forest. Symbiosis 31 (1–3): 85–94.
- Sanmartín, P., Rodríguez, A. & Aguiar, U. (2020) Medium-term field evaluation of several widely used cleaning-restoration techniques applied to algal biofilm formed on a granite-built historical monument. International Biodeterioration & Biodegradation 147: 104870. https://doi.org/10.1016/j.ibiod.2019.104870
- Sophia, M.G., Huszar, V.L. M., Silva, L.H.S., Domingues, C.D., Santos, J.B.O. & Bicudo, C.E.M. (2016) Subaerial eukaryotic algae and cyanobacteria on dripping rocks in the Atlantic Forest of southeast Brazil: composition and abundance. Brazilian Journal of Botany 39 (2): 741–749. https://doi.org/10.1007/s40415-016-0261-3
- Wang, Q.H., Liu, X.D., Li, S.Y., Xiong, Q., Hu, Z.Y. & Liu, G.X. (2020) Cryptic species inside the genus Hariotina (Scenedesmaceae, Sphaeropleales), with descriptions of four new species in this genus. European Journal of Phycology 55 (4): 373–383. https://doi.org/10.1080/09670262.2020.1737968
- Zechman, F.W. (2003) Phylogeny of the Dasycladales (Chlorophyta, Ulvophyceae) based on analyses of RUBISCO large subunit (rbcL) gene sequences. Journal of Phycology 39 (4): 819–827. https://doi.org/10.1046/j.1529-8817.2003.02183.x
- Zhu, H., Li, S.Y., Hu, Z.Y. & Liu, G.X. (2018) Molecular characterization of eukaryotic algal communities in the tropical phyllosphere based on real-time sequencing of the 18S rDNA gene. BMC Plant Biology 18. https://doi.org/10.1186/s12870-018-1588-7
- Zhu, H., Zhao, Z.J., Xia, S., Hu, Z.Y. & Liu, G.X. (2015) Morphological Examination and Phylogenetic Analyses of Phycopeltis spp. (Trentepohliales, Ulvophyceae) from Tropical China. PLOS One 10 (2): e0114936. https://doi.org/10.1371/journal.pone.0114936