Abstract
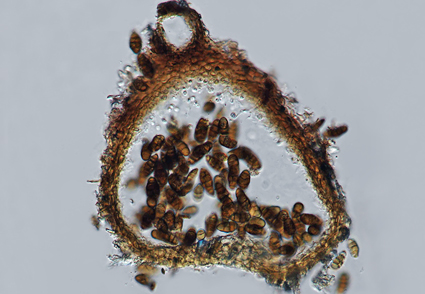
In this study, a saprobic species of Murichromolaenicola (Phaeosphaeriaceae, Dothideomycetes) was collected from dead stems of Chromolaena odorata in northern Thailand. It is introduced as a new species Murichromolaenicola thailandensis sp. nov. based on morphology and multigene phylogeny of combined LSU, SSU, ITS, tef1-α, and rpb2 sequence data. Murichromolaenicola thailandensis is similar to other Murichromolaenicola species by having muriform spores but can be differentiated by its smaller conidia with a gelatinous cap and the number of septa on conidia. Here, we present a detailed description, illustrations and antibacterial potential of this new species. Our findings contribute to the expanding knowledge of fungal diversity in Thailand and highlight the importance of utilizing molecular approaches to accurately identify and classify fungi in Ascomycota.
References
- Ahmed, S.A., Hofmüller, W., Seibold, M., De Hoog, G.S., Harak, H., Tammer, I., Van Diepeningen, A.D. & Behrens‐Baumann, W. (2017) Tintelnotia, a new genus in Phaeosphaeriaceae harbouring agents of cornea and nail infections in humans. Mycoses 60: 244–253. https://doi.org/10.1111/myc.12588
- Alam, S.T., Le, T.A.N., Park, J.S., Kwon, H.C. & Kang, K. (2019) Antimicrobial biophotonic treatment of ampicillin-resistant Pseudomonas aeruginosa with hypericin and ampicillin cotreatment followed by orange light. Pharmaceutics 11: 641. https://doi.org/10.3390/pharmaceutics11120641
- Almeida, C., Mackenzie, T.A., González-Menéndez, V., de Pedro, N., Pérez-Victoria, I., Crespo, G., Martín, J., Genilloud, O., Vicente, F., Cautain, B. & Reyes, F. (2019) Laccaridione C, a bioactive polyketide from the fungus Montagnula sp. Planta Medica International Open 6: e57–e62. https://doi.org/10.1055/a-1032-3346
- Balouiri, M., Sadiki, M. & Ibnsouda, S.K. (2016) Methods for in vitro evaluating antimicrobial activity: A review. Journal of pharmaceutical analysis 6: 71–79. https://doi.org/10.1016/j.jpha.2015.11.005
- Barr, M.E. (1979) A classification of Loculoascomycetes. Mycologia 71: 935–957. https://doi.org/10.1080/00275514.1979.12021099
- Catarino, L., Indjai, B., Duarte, M.C. & Monteiro, F. (2019) Chromolaena odorata invasion in Guinea-Bissau (West Africa): first records and trends of expansion. BioInvasions Records 8: 190–198. https://doi.org/10.3391/bir.2019.8.1.20
- Chethana, K.T., Manawasinghe, I.S., Hurdeal, V.G., Bhunjun, C.S., Appadoo, M.A., Gentekaki, E., Raspé, O., Promputtha, I. & Hyde, K.D. (2021) What are fungal species and how to delineate them? Fungal Diversity 109: 1–25. https://doi.org/10.1007/s13225-021-00483-9
- Cox, J. & Lambert, J. (2013) Microsoft PowerPoint 2013. Microsoft Press, Washington, 481 pp.
- Crous, P.W., Cowan, D.A., Maggs-Kölling, G., Yilmaz, N., Thangavel, R., Wingfield, M.J., Noordeloos, M.E., Dima, B., Brandrud, T.E., Jansen, G.M. & Morozova, O.V. (2021) Fungal Planet description sheets: 1182–1283. Persoonia: Molecular Phylogeny and Evolution of Fungi 46: 313. https://doi.org/10.3767/persoonia.2021.46.11
- Crous, P.W., Luangsa-Ard, J.J., Wingfield, M.J., Carnegie, A.J., Hernández-Restrepo, M., Lombard, L., Roux, J., Barreto, R.W., Baseia, I.G., Cano-Lira, J.F. & Martín, M.P. (2018) Fungal Planet description sheets: 785–867. Persoonia-Molecular Phylogeny and Evolution of Fungi 41: 238–417. https://doi.org/10.3767/persoonia.2018.41.12
- Crous, P.W., Schumacher, R.K., Akulov, A., Thangavel, R., Hernández-Restrepo, M., Carnegie, A.J., Cheewangkoon, R., Wingfield, M.J., Summerell, B.A., Quaedvlieg, W. & Coutinho, T.A. (2019) New and interesting fungi 2. Fungal Systematics and Evolution 3: 57. https://doi.org/10.3114/fuse.2019.03.06
- Dai, D.Q., Phookamsak, R., Wijayawardene, N.N., Li, W.J., Bhat, D.J., Xu, J.C., Taylor, J.E., Hyde, K.D. & Chukeatirote, E. (2017) Bambusicolous fungi. Fungal Diversity 82: 1–105. https://doi.org/10.1007/s13225-016-0367-8
- Devadatha, B., Mehta, N., Wanasinghe, D.N., Baghela, A. & Sarma, V.V. (2019) Vittaliana mangrovei, gen. nov., sp. nov. (Phaeosphaeriaceae) from mangroves near Pondicherry (India), based on morphology and multigene phylogeny. Cryptogamie, Mycologie 40: 117–132. https://doi.org/10.5252/cryptogamiemycologie2019v40a7
- Dissanayake, L.S., Marasinghe, D.S., Thambugala, K.M. & Kang, J.C. (2022) Neostagonosporella bambusicola sp. nov. (Phaeosphaeriaceae, Pleosporales) from bamboo in China. Phytotaxa 573: 262–274. https://doi.org/10.11646/phytotaxa.573.2.6
- Doilom, M., Dissanayake, A.J., Wanasinghe, D.N., Boonmee, S., Liu, J.K., Bhat, D.J., Taylor, J.E., Bahkali, A.H., McKenzie, E.H. & Hyde, K.D. (2017) Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Diversity 82: 107–182. https://doi.org/10.1007/s13225-016-0368-7
- Gafforov, Y., Phookamsak, R., Jiang, H.B., Wanasinghe, D.N. & Juliev, M. (2019) Ophiobolus hydei sp. nov. (Phaeosphaeriaceae, Ascomycota) from Cirsium and Phlomoides in Uzbekistan. Botany 97: 671–680. https://doi.org/10.1139/cjb-2019-0118
- Goodall, J.M. & Erasmus, D.J. (1996) Review of the status and integrated control of the invasive alien weed, Chromolaena odorata, in South Africa. Agriculture, ecosystems & environment 56: 151–164. https://doi.org/10.1016/0167-8809(95)00647-8
- Hall, T.A. (1990) BioEdit: a user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [http://wwwmbio-ncsuedu/bioedit/bioedithtml]
- Hernandez-Restrepo, M., Schumacher, R.K., Wingfield, M.J., Ahmad, I., Cai, L., Duong, T.A., Edwards, J., Gene, J., Groenewald, J.Z., Jabeen, S. & Khalid, A.N. (2016) Fungal systematics and evolution: FUSE 2. Sydowia 68: 193–230.
- Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H.C., Sarma, V.V., Boonmee, S., Lücking, R., Bhat, D.J., Liu, N.G. & Tennakoon, D.S. (2020) Refined families of dothideomycetes: Dothideomycetidae and pleosporomycetidae. Mycosphere 11: 1553–2107. https://doi.org/10.5943/mycosphere/11/1/13
- Hridhya, K.V. & Kulandhaivel, M. (2017) Antimicrobial activity of Chromolaena odorata against selected pyogenic pathogens. International Journal of Pharmacognosy and Phytochemical Research 9: 1001–1007. https://doi.org/10.25258/phyto.v9i07.11171
- Hu, G. & Zhang, Z. (2013) Allelopathic effects of Chromolaena odorata on native and non-native invasive herbs. Journal of Food, Agriculture & Environment 11: 878–882.
- Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., McKenzie, E.H., Jones, E.G., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q. & Abdel-Aziz, F.A. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal diversity 80: 1–270. https://doi.org/101007/s13225-016-0373-x
- Hyde, K.D., Norphanphoun, C., Abreu, V.P., Bazzicalupo, A., Thilini Chethana, K.W., Clericuzio, M., Dayarathne, M.C., Dissanayake, A.J., Ekanayaka, A.H., He, M.Q. & Hongsanan, S. (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87: 1–235. https://doi.org/10.1007/s13225-017-0391-3
- Hyde, K.D., Tennakoon, D.S., Jeewon, R., Bhat, D.J., Maharachchikumbura, S.S., Rossi, W., Leonardi, M., Lee, H.B., Mun, H.Y., Houbraken, J. & Nguyen, T.T. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. https://doi.org/10.1007/s13225-019-00429-2
- Jayasiri, S.C., Hyde, K.D., Ariyawansa, H.A., Bhat, J., Buyck, B., Cai, L., Dai, Y.C., Abd-Elsalam, K.A., Ertz, D., Hidayat, I. & Jeewon, R. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal diversity 74: 3–18. https://doi.org/10.1007/s13225-015-0351-8
- Karunarathna, A., Papizadeh, M., Senanayake, I.C., Jeewon, R., Phookamsak, R., Goonasekara, I.D., Wanasinghe, D.N., Wijayawardene, N.N., Amoozegar, M.A., Shahzadeh Fazeli, S.A. & Camporesi, E. (2017) Novel fungal species of Phaeosphaeriaceae with an asexual/sexual morph connection. Mycosphere 8: 1818–1834. https://doi.org/10.5943/mycosphere/8/10/8
- Karunarathna, A., Phookamsak, R., Jayawardena, R.S., Hyde, K.D. & Kuo, C.H. (2020) Kwanghwana miscanthi Karun., CH Kuo & KD Hyde, gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Miscanthus floridulus (Labill.) Warb. ex K. Schum. & Lauterb.(Poaceae). Cryptogamie, Mycologie 41: 119–132. https://doi.org/10.5252/cryptogamiemycologie2020v41a6
- Lemmon, A.R., Brown, J.M., Stanger-Hall, K. & Lemmon, E.M. (2009) The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Systematic biology 58: 130–145. https://doi.org/10.1093/sysbio/syp017
- Li, J.F., Phookamsak, R., Jeewon, R., Bhat, D.J., Mapook, A., Camporesi, E., Shang, Q.J., Chukeatirote, E., Bahkali, A.H. & Hyde, K.D. (2017) Molecular taxonomy and morphological characterization reveal new species and new host records of Torula species (Torulaceae, Pleosporales). Mycological progress 16: 447–461. https://doi.org/10.1007/s11557-017-1292-2
- Li, W.J., Bhat, D.J., Camporesi, E., Tian, Q., Wijayawardene, N.N., Dai, D.Q., Phookamsak, R., Chomnunti, P., Bahkali, A.H. & Hyde, K.D. (2015) New asexual morph taxa in Phaeosphaeriaceae. Mycosphere 6: 681–708. https://doi.org/10.5943/mycosphere/6/6/5
- Li, W.J., McKenzie, E.H., Liu, J.K., Bhat, D.J., Dai, D.Q., Camporesi, E., Tian, Q., Maharachchikumbura, S.S., Luo, Z.L., Shang, Q.J. & Zhang, J.F. (2020) Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Diversity 100: 279–801. https://doi.org/10.1007/s13225-020-00440-y
- Liu, J.K., Hyde, K.D., Jones, E.G., Ariyawansa, H.A., Bhat, D.J., Boonmee, S., Maharachchikumbura, S.S., McKenzie, E.H., Phookamsak, R., Phukhamsakda, C. & Shenoy, B.D. (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal diversity 72: 1–197.
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular biology and evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Maharachchikumbura, S.S., Ariyawansa, H.A., Wanasinghe, D.N., Dayarathne, M.C., Al-Saady, N.A. & Al-Sadi, A.M. (2019) Phylogenetic classification and generic delineation of Hydeomyces desertipleosporoides gen. et sp. nov., (Phaeosphaeriaceae) from Jebel Akhdar Mountain in Oman. Phytotaxa 391: 28–38. https://doi.org/10.11646/phytotaxa.391.1.2
- Mapook, A., Hyde, K.D., Dai, D.Q., Li, J., Jones, E.G., Bahkali, A.H. & Boonmee, S. (2016) Muyocopronales, ord. nov. (Dothideomycetes, Ascomycota) and a reappraisal of Muyocopron species from northern Thailand. Phytotaxa 265: 225–237. https://doi.org/10.11646/phytotaxa.265.3.3
- Mapook, A., Hyde, K.D., McKenzie, E.H., Jones, E.G., Bhat, D.J., Jeewon, R., Stadler, M., Samarakoon, M.C., Malaithong, M., Tanunchai, B. & Buscot, F. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Diversity 101: 1–175. https://doi.org/10.1007/s13225-020-00444-8
- Mathew, M., Anil, A., Carlin, V.V., Raveendran, R., Ruban, N. & Jannet, V.J. (2022) An updated research on the antimicrobial properties of Chromolaena odorata (L). leaf and flower extracts against wound promoting pathogens: A comparative and combinatorial in vitro/in silico approach. Medicinal Plants-International Journal of Phytomedicines and Related Industries 14: 129–143. https://doi.org/10.5958/0975-6892.2022.00014.4
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE), Presented at the 2010 Gateway Computing Environments Workshop (GCE), IEEE, New Orleans, LA, USA: 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Miyake, I. (1909) Studies on the parasitic fungi of rice in Japan. Botanical Magazine Tokyo 23: 85–97. https://doi.org/1015281/jplantres188723266_85
- Muniappan, R., Reddy, G.V.P. & Lai, P.Y. (2005) Distribution and biological control of Chromolaena odorata. In: Inderjit, S. (Ed.) Invasive plants: ecological and agricultural aspects. Birkhäuser Basel, pp. 223–233. https://doi.org/10.1007/3-7643-7380-6_14
- Phookamsak, R., Hyde, K.D., Jeewon, R., Bhat, D.J., Jones, E.G., Maharachchikumbura, S.S., Raspé, O., Karunarathna, S.C., Wanasinghe, D.N., Hongsanan, S. & Doilom, M. (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal diversity 95: 1–273. https://doi.org/101007/s13225-019-00421-w
- Phookamsak, R., Liu, J.K., McKenzie, E.H., Manamgoda, D.S., Ariyawansa, H., Thambugala, K.M., Dai, D.Q., Camporesi, E., Chukeatirote, E., Wijayawardene, N.N. & Bahkali, A.H. (2014) Revision of Phaeosphaeriaceae. Fungal Diversity 68: 159–238. https://doi.org/10.1007/s13225-014-0308-3
- Phookamsak, R., Wanasinghe, D.N., Hongsanan, S., Phukhamsakda, C., Huang, S.K., Tennakoon, D.S., Norphanphoun, C., Camporesi, E., Bulgakov, T.S., Promputtha, I. & Mortimer, P.E. (2017) Towards a natural classification of Ophiobolus and Ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales). Fungal Diversity 87: 299–339. https://doi.org/10.1007/s13225-017-0393-1
- Phukhamsakda, C., Ariyawansa, H.A., Phookamsak, R., Chomnunti, P., Bulgakov, T.S., Yange, J.B., Bhat, D.J., Bahkalih, A.H. & Hyde, K.D. (2015) Muriphaeosphaeria galatellae gen. et sp. nov. in Phaeosphaeriaceae (Pleosporales). Phytotaxa 227: 55–65. https://doi.org/10.11646/phytotaxa.227.1.6
- Phukhamsakda, C., McKenzie, E.H., Phillips, A.J., Gareth Jones, E.B., Jayarama Bhat, D., Stadler, M., Bhunjun, C.S., Wanasinghe, D.N., Thongbai, B., Camporesi, E. & Ertz, D. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal diversity 102: 1–203. https://doi.org/10.1007/s13225-020-00448-4
- Rambaut, A. (2012) FigTree v.1.40. University of Oxford. https://doi.org/10.1111/j.1548-1352.2012.01262.x
- Rannala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of molecular evolution 43: 304–311. https://doi.org/10.1007/BF02338839
- Rehner, S. (2001) Primers for Elongation Factor 1–alpha (EF1–alpha). Insect Biocontrol Laboratory: USDA, ARS, PSI.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 32: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N. & Bundhun, D. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. http://doi.org/105943/mycosphere/11/1/20
- Sinha, A., Nath, A., Lahkar, B.P., Brahma, N., Sarma, H.K. & Swargowari, A. (2022) Understanding the efficacy of different techniques to manage Chromolaena odorata L., an Invasive Alien Plant in the sub-Himalayan tall grasslands: Toward grassland recovery. Ecological Engineering 179: 106618. https://doi.org/10.1016/j.ecoleng.2022.106618
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post–analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Tennakoon, D.S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., Camporesi, E. & Promputtha, I. (2016) Taxonomy and phylogeny of Juncaceicola gen. nov. (Phaeosphaeriaceae, Pleosporinae, Pleosporales). Cryptogamie, Mycologie 37: 135–156. https://doi.org/10.7872/crym/v37.iss2.2016.135
- Tennakoon, D.S., Thambugala, K.M., Wanasinghe, D.N., Gentekaki, E., Promputtha, I., Kuo, C.H. & Hyde, K.D. (2020) Additions to Phaeosphaeriaceae (Pleosporales): Elongaticollum gen. nov., Ophiosphaerella taiwanensis sp. nov., Phaeosphaeriopsis beaucarneae sp. nov. and a new host record of Neosetophoma poaceicola from Musaceae. MycoKeys 70: 59. https://doi.org/10.3897/mycokeys.70.53674.figure2
- Thambugala, K.M., Wanasinghe, D.N., Phillips, A.J.L., Camporesi, E., Bulgakov, T.S., Phukhamsakda, C., Ariyawansa, H.A., Goonasekara, I.D., Phookamsak, R., Dissanayake, A. & Tennakoon, D.S. (2017) Mycosphere notes 1–50: grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 8: 697–796. https://doi.org/10.5943/mycosphere/8/4/13
- Thophon, S.H.S., Waranusantigul, P., Kangwanrangsan, N. & Krajangsang, S. (2016) Antimicrobial activity of Chromolaena odorata extracts against bacterial human skin infections. Modern Applied Science 10: 159. https://doi.org/10.5539/mas.v10n2p159
- Tibpromma, S., Hyde, K.D., McKenzie, E.H., Bhat, D.J., Phillips, A.J., Wanasinghe, D.N., Samarakoon, M.C., Jayawardena, R.S., Dissanayake, A.J., Tennakoon, D.S. & Doilom, M. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal diversity 93: 1–160. https://doi.org/10.1007/s13225-018-0408-6
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- Vital, P.G. & Rivera, W.L. (2009) Antimicrobial activity and cytotoxicity of Chromolaena odorata (L. f.) King and Robinson and Uncaria perrottetii (A. Rich) Merr. Extracts. Journal of medicinal plants Research 3: 511–518. https://doi.org/10.5897/JMPR.9000160
- Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Groenewald, J.Z., Cardinali, G., Houbraken, J. & Boekhout, T. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in mycology 91: 23–36. https://doi.org/10.1016/j.simyco.2018.05.001
- Wanasinghe, D.N., Hyde, K.D., Jeewon, R., Crous, P.W., Wijayawardene, N.N., Jones, E.B.G., Bhat, D.J., Phillips, A.J., Groenewald, J.Z., Dayarathne, M.C. & Phukhamsakda, C. (2017) Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Studies in Mycology 87: 207–256. https://doi.org/10.1016/j.simyco.2017.08.001
- Wanasinghe, D.N., Phukhamsakda, C., Hyde, K.D., Jeewon, R., Lee, H.B., Gareth Jones, E.B., Tibpromma, S., Tennakoon, D.S., Dissanayake, A.J., Jayasiri, S.C. & Gafforov, Y. (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal diversity 89: 1–236. https://doi.org/10.1007/s13225-018-0395-7
- White, T.J., Bruns, T., Lee, S.J.W.T. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sánchez-García, M., Goto, B.T. & Magurno, F. (2022) Outline of Fungi and fungus-like taxa-2021. Mycosphere 13: 53–453. https://doi.org/105943/mycosphere/13/1/2
- Wijayawardene, N.N., Song, Y., Bhat, D.J., McKenzie, E.H., Chukeatirote, E., Wang, Y. & Hyde, K.D. (2013) Wojnowicia viburni, sp. nov., from China and its phylogenetic placement. Sydowia 65: 129–138.
- Yang, C.L., Xu, X.L., Wanasinghe, D.N., Jeewon, R., Phookamsak, R., Liu, Y.G., Liu, L.J. & Hyde, K.D. (2019) Neostagonosporella sichuanensis gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Phyllostachys heteroclada (Poaceae) from Sichuan Province, China. MycoKeys 46: 119. https://doi.org/10.3897/mycokeys.46.32458
- Zachariades, C., Janse van Rensburg, S. & Witt, A. (2010) Recent spread and new records of Chromolaena odorata in Africa. In: Proceedings of the eighth workshop on biological control and management of Chromolaena odorata and other Eupatorieae. Nairobi ARC-PPRI, Pretoria, pp. 1–2.
- Zhang, Y., Schoch, C.L., Fournier, J., Crous, P.W., De Gruyter, J., Woudenberg, J.H.C., Hirayama, K., Tanaka, K., Pointing, S.B., Spatafora, J.W. & Hyde, K.D. (2009) Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85–102. https://doi.org/10.3114/sim.2009.64.04
- Zhang, J.F., Liu, J.K., Jeewon, R., Wanasinghe, D.N. & Liu, Z.Y. (2019) Fungi from Asian Karst formations III. Molecular and morphological characterization reveal new taxa in Phaeosphaeriaceae. Mycosphere 10: 202–220. https://doi.org/10.5943/mycosphere/10/1/3
- Zhao, Z.Z., Zhao, K., Chen, H.P., Bai, X., Zhang, L. & Liu, J.K. (2018) Terpenoids from the mushroom-associated fungus Montagnula donacina. Phytochemistry 147: 21–29. https://doi.org/10.1016/j.phytochem.2017.12.015
- Zhaxybayeva, O. & Gogarten, J.P. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC genomics 3: 1–15. https://doi.org/10.1186/1471-2164-3-4