Abstract
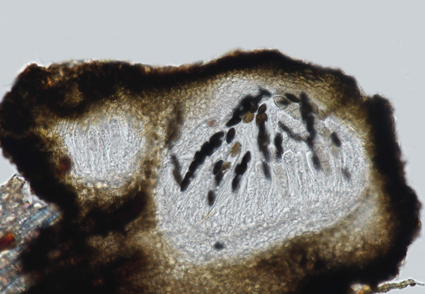
Dothiorella has cosmopolitan species and a wide range of hosts as endophytes, saprobes, and pathogens. Many species have been introduced solely based on their host association, resulting in misidentification. This study describes a novel species, Dothiorella saprophytica sp. nov., a sexual morph of Dothiorella, from Chiang Mai, Thailand using multi-locus phylogeny (ITS + tef1-α + β-tub) and morphological characters. Detailed morphology and multi-locus phylogeny are provided to give more insights into the diversity and taxonomy of this genus. Dothiorella saprophytica is characterized by its sexual morph with pseudothecial and immersed ascomata, peridium textura angularis, hyaline, thin-walled, unbranched, and septate pseudoparaphyses, clavate and bitunicate asci, and ovoid to sub-clavate, 0–1-septate ascospores. Introducing new fungal species, such as D. saprophytica sp. nov., adds to our understanding of biodiversity and the ecology of fungi and provides opportunities for further research.
References
- Abdollahzadeh, J., Javadi, A., Zare, R. & Phillips, A.J.L. (2014) A phylogenetic study of Dothiorella and Spencermartinsia species associated with woody plants in Iran, New Zealand, Portugal and Spain. Persoonia 32: 1–2. https://doi.org/10.3767/003158514X678606
- Burgess, T.I., Barber, P.A., Mohali, S., Pegg, G., de Beer, W. & Wingfield, M.J. (2006) Three new Lasiodiplodia spp. from the tropics, recognized based on DNA sequence comparisons and morphology. Mycologia 98: 423–435. https://doi.org/10.1080/15572536.2006.11832677
- Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
- Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. https://doi.org/10.1080/00275514.1999.12061051
- Chen, S., Morgan, D.P., Hasey, J.K., Anderson, K. & Michailides, T.J. (2014) Phylogeny, morphology, distribution, and pathogenicity of Botryosphaeriaceae and Diaporthaceae from English walnut in California. Plant Disease 98: 636–652. https://doi.org/10.1094/PDIS-07-13-0706-RE
- Chernomor, O., Von Haeseler, A. & Minh, B.Q. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. https://doi.org/10.1093/sysbio/syw037
- Chethana, K.T., Manawasinghe, I.S., Hurdeal, V.G., Bhunjun, C.S., Appadoo, M.A., Gentekaki, E., Raspé, O., Promputtha, I. & Hyde, K.D. (2021) What are fungal species and how to delineate them? Fungal Diversity 109: 1–25. https://doi.org/10.1007/s13225-021-00483-9
- Crous, P.W. & Palm, M.E. (1999) Reassessment of the anamorph genera Botryodiplodia, Dothiorella and Fusicoccum. Sydowia 51: 167–75.
- Darriba, D., Posada, D., Kozlov, A.M., Stamatakis, A., Morel, B. & Flouri, T. (2020) ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37: 291–294. https://doi.org/10.1093/molbev/msz189
- De Wet, J., Slippers, B., Preisig, O., Wingfield, B.D., Tsopelas, P. & Wingfield, M.J. (2009) Molecular and morphological characterization of Dothiorella casuarini sp. nov. and other Botryosphaeriaceae with diplodia-like conidia. Mycologia 101: 503–11. https://doi.org/10.3852/07-180
- Dissanayake, A.J., Camporesi, E., Hyde, K.D., Phillips, A.J., Fu, C.Y., Yan, J.Y. & Li, X.H. (2016) Dothiorella species associated with woody hosts in Italy. Mycosphere 7 (1): 51–63. https://doi.org/10.5943/mycosphere/7/1/6
- Doll, D.A., Rolshausen, P.E., Pouzoulet, J. & Michailides, T.J. (2015) First report of Dothiorella iberica causing trunk and scaffold cankers of almond in California. Plant Disease 99: 1185. https://doi.org/10.1094/PDIS-11-14-1233-PDN
- Glass, N.L. & Donaldson, G.C. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental microbiology 61: 1323–1330. https://doi.org/10.1128/aem.61.4.1323-1330.1995
- Phillips, A.J., Alves, A., Pennycook, S.R., Johnston, P.R., Ramaley, A., Akulov, A. & Crous, P.W. (2008) Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia-Molecular Phylogeny and Evolution of Fungi: 129–55. https://doi.org/10.3767/003158508X340742
- Hoang, D.T., Chernomor, O., Von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281
- Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H., Sarma, V.V., Lücking, R., Boonmee, S., Bhat, J.D., Liu, N.G. & Tennakoon, D.S. (2020) Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105: 17–318. https://doi.org/10.1007/s13225-020-00462-6
- Hyde, K.D., de Silva, N.I., Jeewon, R., Bhat, D.J., Phookamsak, R., Doilom, M., Boonmee, S., Jayawardena, R.S., Maharachchikumbura, S.S.N., Senanayake, I.C., Manawasinghe, I.S., Liu, N.G., Abeywickrama, P.D., Chaiwan, N., Karunarathna, A., Pem, D., Lin C.G., Sysouphanthong, P., Luo, Z.L., Wei, D.P., Wanasinghe, D.N., Norphanphoun, C., Tennako, D.S., Samarakoon, M.C., Jayasiri, S.C., Jiang, H.B., Zeng, X.Y., Li J.F., Wijesinghe, S. N., Devadatha, B., Goonasekara, I.D., Brahmanage, R.S., Yang, E.F., Aluthmuhandiram, J.V.S., Dayarathne, M.C., Marasinghe, D.S., Li, W.J., Dissanayake, L.S., Dong, W., Huanraluek, N., Lumyong, S., Liu, J.K., Karunarathna, S.C., Jones, E.B.G., Al-Sadi, A.M., Xu, J.C., Harishchandra, D. & Sarma, V.V. (2020) AJOM new records and collections of fungi: 1-100 Asian Journal of Mycology 3: 22–294. https://doi.org/10.5943/ajom/3/1/3
- Hyde, K.D., Xu, J., Rapior, S., Jeewon, R., Lumyong, S., Niego, A.G., Abeywickrama, P.D., Aluthmuhandiram, J.V., Brahamanage, R.S., Brooks, S. & Chaiyasen, A. (2019) The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Diversity 97: 1–36. https://doi.org/10.1007/s13225-019-00430-9
- Jayawardena, R.S., Hyde, K.D., McKenzie, E.H., Jeewon, R., Phillips, A.J., Perera, R.H., de Silva, N.I., Maharachchikumburua, S.S., Samarakoon, M.C., Ekanayake, A.H. & Tennakoon, D.S. (2019) One stop shop III: taxonomic update with molecular phylogeny for important phytopathogenic genera: 51-75 (2019) Fungal Diversity 98: 77–160. https://doi.org/10.1007/s13225-019-00433-6
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K., Von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010
- Lawrence, D.P., Hand, F.P., Gubler, W.D. & Trouillas, F.P. (2017) Botryosphaeriaceae species associated with dieback and canker disease of bay laurel in northern California with the description of Dothiorella californica sp. nov. Fungal Biology 121: 347–360. https://doi.org/10.1016/j.funbio.2016.09.005
- Li, W.L., Liang, R.R., Dissanayake, A.J. & Liu, J.K. (2023) Botryosphaerialean fungi associated with woody oil plants cultivated in Sichuan Province, China. MycoKeys 97: 71. https://doi.org/10.3897/mycokeys.97.103118
- Li, W.L., Liang, R.R., Dissanayake, A.J. & Liu, J.K. (2023) Morphology and molecular analyses reveal three new species of Botryosphaeriales isolated from diseased plant branches in China. MycoKeys 97: 1. https://doi.org/10.3897/mycokeys.97.102653
- Liu, J.K., Phookamsak, R., Doilom, M., Wikee, S., Li, Y.M., Ariyawansa, H., Boonmee, S., Chomnunti, P., Dai, D.Q., Bhat, J.D. & Romero, A.I. (2012) Towards a natural classification of Botryosphaeriales. Fungal Diversity 57: 149–210. https://doi.org/10.1007/s13225-012-0207-4
- Luque, J., Martos, S. & Phillips, A.J. (2005) Botryosphaeria viticola sp. nov. on grapevines: a new species with a Dothiorella anamorph. Mycologia 97: 1111–21. https://doi.org/10.1080/15572536.2006.11832759
- Lynch, S.C., Eskalen, A., Zambino, P.J., Mayorquin, J.S. & Wang, D.H. (2013) Identification and pathogenicity of Botryosphaeriaceae species associated with coast live oak (Quercus agrifolia) decline in southern California. Mycologia 105: 125–140. https://doi.org/10.3852/12-047
- Lynch, S.C., Zambino, P.J., Scott, T.A. & Eskalen, A. (2014) Occurrence, incidence and associations among fungal pathogens and Agrilus auroguttatus, and their roles in Quercus agrifolia decline in California. Forest Pathology 44: 62–74. https://doi.org/10.1111/efp.12070
- Maharachchikumbura, S.S., Chen, Y., Ariyawansa, H.A., Hyde, K.D., Haelewaters, D., Perera, R.H., Samarakoon, M.C., Wanasinghe, D.N., Bustamante, D.E., Liu, J.K. & Lawrence, D.P. (2021) Integrative approaches for species delimitation in Ascomycota. Fungal Diversity 109: 155–179. https://doi.org/10.1007/s13225-021-00486-6
- Mapook, A., Hyde, K.D., Hassan, K., Kemkuignou, B.M., Čmoková, A., Surup, F., Kuhnert, E., Paomephan, P., Cheng, T., de Hoog, S. & Song, Y. (2022) Ten decadal advances in fungal biology leading towards human well-being. Fungal Diversity 116: 547–614. https://doi.org/10.1007/s13225-022-00510-3
- Nguyen, L.T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. https://doi.org/10.1093/molbev/msu300
- Pan, M., Tian, C.M. & Fan, X.L. (2021) Two fungal species associated with canker disease of Jujube tree in China. Mycoasia 3: 1–21. https://doi.org/10.59265/mycoasia.2021-03
- Phillips, A.J.L., Hyde, K.D., Alves, A. & Liu, J.K. (2019) Families in Botryosphaeriales: a phylogenetic, morphological and evolutionary perspective. Fungal Diversity 94: 1–22. https://doi.org/10.1007/s13225-018-0416-6
- Phillips, A.J., Alves, A., Abdollahzadeh, J., Slippers, B., Wingfield, M.J., Groenewald, J.Z. & Crous, P.W. (2013) The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology: 51–167. https://doi.org/10.3114/sim0021
- Phillips, A., Alves, A., Correia, A. & Luque, J. (2005) Two new species of Botryosphaeria with brown, 1-septate ascospores and Dothiorella anamorphs. Mycologia 97: 513. https://doi.org/10.3852/mycologia.97.2.513
- Phookamsak, R., Hyde, K.D., Jeewon, R., Bhat, D.J., Jones, E.G., Maharachchikumbura, S.S., Raspé, O., Karunarathna, S.C., Wanasinghe, D.N., Hongsanan, S. & Doilom, M. (2019) Fungal diversity notes 929-1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95: 1–273. https://doi.org/10.1007/s13225-019-00421-w
- Piel, W., Donoghue, M. & Sanderson, M. (2002) TreeBASE: a database of phylogenetic knowledge. In Proceedings of 2nd International Workshop of Species 2000 (Research Report for the National Institute of Environmental Studies, R-171–2002) Tsukuba, Japan.
- Rathnayaka, A.R., Chethana, K.T., Phillips, A.J. & Jones, E.G. (2022) Two new species of Botryosphaeriaceae (Botryosphaeriales) and new host/geographical records. Phytotaxa 564 (1): 8–38. https://doi.org/10.11646/phytotaxa.564.1.2
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–42. https://doi.org/10.1093/sysbio/sys029
- Saccardo, P.A. (1880) Conspectus generum fungorum Italiae inferorium. Michelia 2: 1–38.
- Sayers, E.W., Bolton, E.E., Brister, J.R., Canese, K., Chan, J., Comeau, D.C., Farrell, C.M., Feldgarden, M., Fine, A.M., Funk, K. & Hatcher, E. (2023) Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Research 51: D29. https://doi.org/10.1093/nar/gkac1032
- Sohrabi, M., Mohammadi, H., León, M., Armengol, J. & Banihashemi, Z. (2020) Fungal pathogens associated with branch and trunk cankers of nut crops in Iran. European Journal of Plant Pathology 157: 327–351. https://doi.org/10.1007/s10658-020-01996-w
- Úrbez-Torres, J.R. & Gubler, W.D. (2009) Pathogenicity of Botryosphaeriaceae species isolated from grapevine cankers in California. Plant Disease 93: 584–592. https://doi.org/10.1094/PDIS-93-6-0584
- Úrbez-Torres, J.R., Peduto, F., Striegler, R.K., Urrea-Romero, K.E., Rupe, J.C., Cartwright, R.D. & Gubler, W.D. (2012) Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Diversity 52: 169–189. https://doi.org/10.1007/s13225-011-0110-4
- Vaidya, G., Lohman, D.J. & Meier, R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27 (2): 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
- White, T.J., Bruns, T., Lee, S.J. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A. (Ed.) PCR protocols: a guide to methods and applications, Vol. 18, No. 1. Academic Press, Michigan, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sánchez-García, M., Goto, B.T., Saxena, R.K., Erdoğdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., Błaszkowski, J., Boonyuen, N., da Silva, G.A., de Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Mešić, A., Monteiro, J.S., Oehl, F., Pawłowska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro, I., Tang, L.Z., Stadler, M., Yurkov, A. & Thines, A. (2022) Outline of Fungi and fungus-like taxa - 2021. Mycosphere 13: 53–453. https://doi.org/10.5943/mycosphere/13/1/2
- Xiao, X.E., Wang, W., Crous, P.W., Wang, H.K., Jiao, C., Huang, F., Pu, Z.X., Zhu, Z.R. & Li, H.Y. (2021) Species of Botryosphaeriaceae associated with citrus branch diseases in China. Persoonia-Molecular Phylogeny and Evolution of Fungi 47: 106–135. https://doi.org/10.3767/persoonia.2021.47.03
- Yang, T., Groenewald, J.Z., Cheewangkoon, R., Jami, F., Abdollahzadeh, J., Lombard, L. & Crous, P.W. (2017) Families, genera, and species of Botryosphaeriales. Fungal Biology 121: 322–46. https://doi.org/10.1016/j.funbio.2016.11.001
- Zhang, W., Groenewald, J.Z., Lombard, L., Schumacher, R.K., Phillips, A.J. & Crous, P.W. (2021) Evaluating species in Botryosphaeriales. Persoonia-Molecular Phylogeny and Evolution of Fungi 46: 63–115. https://doi.org/10.3767/persoonia.2021.46.03