Abstract
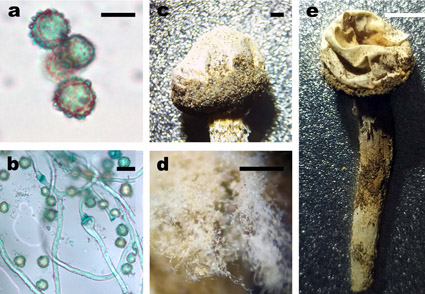
We described a new Tulostoma species based on morphological and molecular data. Its characteristic features are creamy-brownish to grayish white fruiting bodies, spore-sac of small size, stem slender, gleba orange brown, spores are large, subglobose, regularly verrucose, up to 10.6 μm. Warts are large, 6–7 in number on the visible side of spore, up to 0.9 μm length, more or less isolated or merging into groups with small warts or forming ridge-like anastomoses at the base visible on SEM. It grows in scattered shrub communities with Caragana pygmaea, on dry sandy soil in south of Siberia. Based on the sequences of internal transcribed spacer (ITS1-5.8S-ITS2 = ITS) and D1/D2 domain of large subunit (28S) of nuc ribosomal DNA, we reconstructed a phylogeny of the genus Tulostoma. We present the detailed description of new species, color photos of fruiting bodies, light microscopy of anatomical characters, and analysis of morphological features in comparison with related species. To differentiate species of the genus Tulostoma necessary to use scanning electron microscopy. Therefore, we present photographs of spores of a new species under SEM. The DNA sequences obtained by us can also be used for identification of new species.
References
Altschul, S., Gish, W., Miller, W., Myers, E. & Lipman, D. (1990) Basic local alignment search tool. Journal of Molecular Biology 215 (3): 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Binder, M. & Bresinsky, A. (2002) Derivation of a polymorphic lineage of Gasteromycetes from boletoid ancestors. Mycologia 94 (1): 85–98. https://doi.org/10.1080/15572536.2003.11833251
El Kholfy, S., Nmichi, A., Ouabbou, A., Ajana, M., Belahbib, N., Touhami, A.O., Benkirane, R. & Douira, A. (2017) Study of two fungal species of Tulostoma genus encountered for the first time in Morocco: Tulostoma melanocyclum Bres. and Tulostoma kotlabae Pouzar. International Journal of Agriculture Environment and Biotechnology (IJEAB) 2 (1): 235–239. https://doi.org/10.22161/ijeab/2.1.30
Felsenstein, J. (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17: 368–376. https://doi.org/10.1007/BF01734359
Hernández Caffot, M.L., Domínguez, L.S., Hosaka, K. & Crespo, E.M. (2011) Tulostoma domingueziae sp. nov. from Polylepis australis woodlands in Cordoba Mountains, central Argentina. Mycologia 103 (5): 1047–1054. https://doi.org/10.3852/10-266
Hibbett, D.S. & Binder, M. (2002) Evolution of complex fruiting-body morphologies in Homobasidiomycetes. Proceedings of the Royal Society B: Biological Sciences 269: 1963–1969. https://doi.org/10.1098/rspb.2002.2123
Hussain, Sh., Yousaf, N., Afshan, N.U.S., Niazi, A.R., Ahmad, H. & Khalid, A.N. (2016) Tulostoma ahmadii sp. nov. and T. squamosum from Pakistan. Turkish Journal of Botany 40 (2): art. 10. https://doi.org/10.3906/bot-1501-9
Gardes, M. & Bruns, T.D. (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
GBIF (2023) Tulostoma Pers., 1794 in GBIF Secretariat. GBIF Backbone Taxonomy. Checklist dataset https://doi.org/10.15468/39omei accessed via GBIF.org on 03 March 2023.
Gorbunova, I.A. (2003) Macromycetes of the Ukok Plateau (Gorny Altai). Mikologiya I Fitopatologiya 37 (1): 42–49. [in Russian]
Jeppson, M., Altés, A., Moreno, G., Nilsson, R.H., Loarce, Y., de Bustos, A. & Larsson, E. (2017) Unexpected high species diversity among European stalked puffballs – a contribution to the phylogeny and taxonomy of the genus Tulostoma (Agaricales). MycoKeys 21: 33–88. https://doi.org/10.3897/mycokeys.21.12176
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. https://doi.org/10.1093/nar/gkf436
Katoh, K. & Toh, H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9 (4): 286–298. https://doi.org/10.1093/bib/bbn013
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. https://doi.org/10.1093/molbev/msy096
Miller, S.L. & Gongloff, A. (2021) Fairy rings, associated fungi, and assessment of their distribution across environmental variables using GIS. Fungal Ecology 50: 101040. https://doi.org/10.1016/j.funeco.2021.101040
Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32 (1): 268–274. https://doi.org/10.1093/molbev/msu300
Niazi, A.R., Ghafoor, A., Afshan, NuS. & Moreno, G. (2022) Tulostoma loonbanglaense: A new species from Azad Jammu and Kashmir using light and scanning electron microscopy and DNA barcoding technique. Microscopy Research and Technique 85 (12): 3720–3725. https://doi.org/10.1002/jemt.24240
Persoon, C.H. (1794) Neuer Versuch einer systematischen Eintheilung der Schwämme. Neues Magazin für die Botanik 1: 63–128.
Rambaut, A. (2018) FigTree v.1.4.4. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 3 March 2023)
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2, efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
Rusevska, K., Calonge, F.D., Karadelev, M. & Martín, M.P. (2019) Fungal DNA barcode (ITS nrDNA) reveals more diversity than expected in Tulostoma from Macedonia. Turkish Journal of Botany 43 (1): 102–115. https://doi.org/10.3906/bot-1804-38
Trifinopoulos, J., Nguyen, L.T., von Haeseler, A. & Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44 (W1): 232–235. https://doi.org/10.1093/nar/gkw256
Varga, T., Krizsán, K., Földi, C., Dima, B., Sánchez-García, M., Sánchez-Ramírez, S., Szöllősi, G.J., Szarkándi, J.G., Papp, V., Albert, L., Andreopoulos, W., Angelini, C., Antonín, V., Barry, K.W., Bougher, N.L., Buchanan, P., Buyck, B., Bense, V., Catcheside, P., Chovatia, M., Cooper, J., Dämon, W., Desjardin, D., Finy, P., Geml, J., Haridas, S., Hughes, K., Justo, A., Karasiński, D., Kautmanova, I., Kiss, B., Kocsubé, S., Kotiranta, H., LaButti, K.M., Lechner, B.E., Liimatainen, K., Lipzen, A., Lukács, Z., Mihaltcheva, S., Morgado, L.N., Niskanen, T., Noordeloos, M.E., Ohm, R.A., Ortiz-Santana, B., Ovrebo, C., Rácz, N., Riley, R., Savchenko, A., Shiryaev, A., Soop, K., Spirin, V., Szebenyi, C., Tomšovský, M., Tulloss, R.E., Uehling, J., Grigoriev, I.V., Vágvölgyi, C., Papp, T., Martin, F.M., Miettinen, O., Hibbett, D.S. & Nagy, L.G. (2019) Megaphylogeny resolves global patterns of mushroom evolution. Nature Ecology & Evolution 3 (4): 668–678. https://doi.org/10.1038/s41559-019-0834-1
Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Wright, J.E. (1987) The genus Tulostoma (Gasteromycetes) – A world monograph. Bibliotheca Mycologica 113: 338.