Abstract
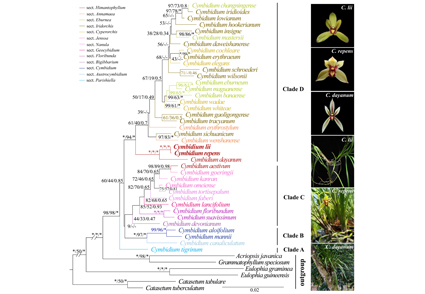
The phylogenetic positions of two little-known species of Cymbidium (Cymbidiinae) with creeping rhizomatous habit, C. lii and C. repens, has previously been unclear. Here, we present a comprehensive molecular phylogenetic study of the genus based on nuclear ribosomal ITS and seven plastid regions. All analyses showed that C. lii and C. repens are sister to C. dayanum, an epiphytic species, with high support. However, many topological conflicts in inter- and intra-sectional relationships were observed in other parts of the trees, supporting existence of hybridization in Cymbidium but not affecting the taxa of interest in this study.
References
Averyanov, L.V., Nong, V.D., Nguyen, K.S., Maisak, T.V., Nguyen, V.C., Phan, Q.T., Nguyen, P.T., Nguyen, T.T. & Truong, B.V. (2016) New species of orchids (Orchidaceae) in the flora of Vietnam. Taiwania 61: 319–354. https://doi.org/10.6165/tai.2016.61.319
Chase, M.W., Cameron, K.M., Freudenstein, J.V., Pridgeon, A.M. & Schuiteman, A. (2015) An updated classification of Orchidaceae. Botanical Journal of the Linnean Society 177: 151–174. https://doi.org/10.1111/boj.12234
Chase, M.W. & Hills, H.H. (1991) Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40: 215–220. https://doi.org/10.2307/1222975
Chen, S.P., Tian, H.Z., Guan, Q.X., Zhai, J.W., Zhang, G.Q., Chen, L.J., Liu, Z.J., Lan, S.R. & Li, M.H. (2019) Molecular systematics of Goodyerinae (Cranichideae, Orchidoideae, Orchidaceae) based on multiple nuclear and plastid regions. Molecular Phylogenetics and Evolution 139: 106542. https://doi.org/10.1016/j.ympev.2019.106542
Chernomor, O., von Haeseler, A. & Minh, B.Q. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. https://doi.org/10.1093/sysbio/syw037
Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. https://doi.org/10.1038/nmeth.2109
De Ré, F.C., Robe, L.J., Wallau, G.L. & Loreto, E.L. (2017) Inferring the phylogenetic position of the Drosophila flavopilosa group: Incongruence within and between mitochondrial and nuclear multilocus datasets. Journal of Zoological Systematics and Evolutionary Research 55: 208–221. https://doi.org/221.10.1111/jzs.12170
Doyle, J.J. & Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
Du Puy, D. & Cribb, P.J. (2007) The genus Cymbidium. Royal Botanic Gardens Kew, UK.
Geuten, K., Smets, E., Schols, P., Yuan, Y.M., Janssens, S., Küpfer, P. & Pyck, N. (2004) Conflicting phylogenies of balsaminoid families and the polytomy in Ericales: combining data in a Bayesian framework. Molecular Phylogenetics and Evolution 31: 711–729. https://doi.org/10.1016/j.ympev.2003.09.014
Guo, Y.Y., Luo, Y.B., Liu, Z.J. & Wang, X.Q. (2015) Reticulate evolution and sea-level fluctuations together drove species diversification of slipper orchids (Paphiopedilum) in South-East Asia. Molecular Ecology 24: 2838–2855. https://doi.org/10.1111/mec. 13189
Huang, M.Z., Liu, Z.J., Yang, G.S. & Yin, J.M. (2017) An unusual new epiphytic species of Cymbidium (Orchidaceae: Epidedroideae) from Hainan, China. Phytotaxa 314: 289–293. https://doi.org/10.11646/phytotaxa.314.2.12
Kanzi, A.M., Trollip, C., Wingfield, M.J., Barnes, I., Van der Nest, M.A. & Wingfield, B.D. (2020) Phylogenomic incongruence in Ceratocystis: a clue to speciation? BMC Genomics 21: 362. https://doi.org/10.1186/s12864-020-6772-0
Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
Li, M.H., Zhang, G.Q., Liu, Z.J. & Lan, S.R. (2016) Subtribal relationships in Cymbidieae (Epidendroideae, Orchidaceae) reveal a new subtribe, Dipodiinae, based on plastid and nuclear coding DNA. Phytotaxa 246: 37–48. https://doi.org/10.11646/phytotaxa.246.1.3
Liu, Z.J. & Chen, S.C. (2004) Cymbidium aestivum, a new species of Orchidaceae from Yunnan. Journal of Wuhan Botanical Research 22: 323–325. [In Chinese]
Liu, Z.J., Chen, S.C. & Ru, Z.Z. (2006) The genus Cymbidium in China. Science Press, Beijing, 360 pp.
Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). IEEE, New Orleans, pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129
Minh, B.Q., Nguyen, M.A.T. & von Haeseler, A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188–1195. https://doi.org/10.1093/molbev/mst024
Minh, B.Q., Schmidt, H.A., Chernomor, O., Schrempf, D., Woodhams, M.D., Von Haeseler, A. & Lanfear, R. (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. https://doi.org/10.1093/molbev/msaa015
Pridgeon, A.M., Cribb, P., Chase, M.W. & Rasmussen, F.N. (2009) Genera orchidacearum, volume 5. Oxford University Press, Oxford, 544 pp.
Reichenbach, H.G. (1869) New garden plants. The Gardeners’ Chronicle 1869: 710.
Richardson, A.O. & Palmer, J.D. (2007) Horizontal gene transfer in plants. Journal of Experimental Botany 58: 1–9. https://doi.org/10.1007/s10142-013-0345-0
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
Russell, A., Samuel, R., Klejna, V., Barfuss, M.H., Rupp, B. & Chase, M.W. (2010) Reticulate evolution in diploid and tetraploid species of Polystachya (Orchidaceae) as shown by plastid DNA sequences and low-copy nuclear genes. Annals of Botany 106: 37–56. https://doi.org/ 10.1093/aob/mcq092
Sharma, S.K., Dkhar, J., Kumaria, S., Tandon, P. & Rao, S.R. (2012) Assessment of phylogenetic inter-relationships in the genus Cymbidium (Orchidaceae) based on internal transcribed spacer region of rDNA. Gene 495: 10–15. https://doi.org/10.1016/j.gene.2011.12.052
Stull, G.W., Pham, K.K., Soltis, P.S. & Soltis, D.E. (2022) Deep reticulation: the long legacy of hybridization in vascular plant evolution. The Plant Journal. [Online Version]. https://doi.org/10.1111/tpj.16142
Swartz, O. (1799) Dionome Epidendri generis. Linn. Nova Acta Regiae Societatis Scientiarum Upsaliensis 6: 61–88.
Swofford, D. (2003) PAUP: phylogenetic analysis using parsimony and other methods, version 4. Sinauer Associates, Sunderland.
Van den Berg, C., Ryan, A., Cribb, P.J. & Chase, M.W. (2002) Molecular phylogenetics of Cymbidium (Orchidaceae: Maxillarieae): sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA and plastid matK. Lindleyana 17: 102–111.
Yang, Z.Y., Ran, J.H. & Wang, X.Q. (2012) Three genome-based phylogeny of Cupressaceae sl: further evidence for the evolution of gymnosperms and Southern Hemisphere biogeography. Molecular Phylogenetics and Evolution 64: 452–470. https://doi.org/10.1186/1471-2148-13-84
Yang, Z.Y., Ran, J.H. & Wang, X.Q. (2012) Three genome-based phylogeny of Cupressaceae s.l.: further evidence for the evolution of gymnosperms and Southern Hemisphere biogeography. Molecular Phylogenetics and Evolution 64: 452–470. https://doi.org/10.1186/1471-2148-13-84
Yang, J.B., Tang, M., Li, H.T., Zhang, Z.R. & Li, D.Z. (2013) Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evolutionary Biology 13: 84. https://doi.org/10.1186/1471-2148-13-84
Yu, W.B., Huang, P.H., Li, D.Z. & Wang, H. (2013) Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS One 8: e74828. https://doi.org/10.1371/journal.pone.0074828
Yukawa, T., Miyoshi, K. & Yokoyama, J. (2002) Molecular phylogeny and character evolution of Cymbidium (Orchidaceae). Bulletin of the National Science Museum, Series B (Botany) 28: 129–139.
Yukawa, T. & Stern, W.L. (2002) Comparative vegetative anatomy and systematics of Cymbidium (Cymbidieae: Orchidaceae). Botanical Journal of the Linnean Society 138: 383–419. https://doi.org/10.1046/j.1095–8339.2002.00038.x
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20: 348–355. https://doi.org/10.1111/1755-0998.13096
Zhang, M.Y., Sun, C.Y., Hao, G., Ye, X.L. & Zhu, G.H. (2002) A preliminary analysis of phylogenetic relationships in Cymbidium (Orchidaceae) based on nrITS sequence data. Journal of Integrative Plant Biology 44: 588–592. https://doi.org/10.3321/j.issn:1672-9072.2002.05.015
Zhang, G.Q., Chen, G.Z., Chen, L.J., Zhai, J.W., Huang, J., Wu, X.Y., Li, M.H., Peng, D.H., Rao, W.H., Liu, Z.J. & Lan, S.R. (2021) Phylogenetic incongruence in Cymbidium orchids. Plant Diversity 43: 452–461. https://doi.org/10.1016/j.pld.2021.08.002