Abstract
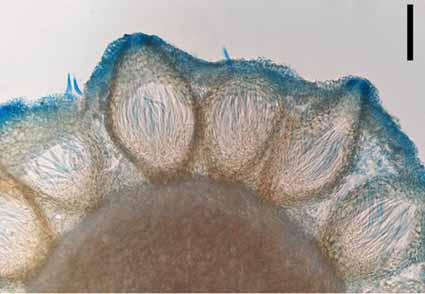
A new species, Ophiocordyceps ovatospora, occurring on termites was collected from Yunnan Province, China, and is described based on morphological and molecular phylogenetic data. In a phylogenetic analysis of a combined dataset of multigene loci comprising the internal transcribed spacer regions (ITS), the small and large subunit of the ribosomal DNA (nrSSU and nrLSU), partial sequences of elongation factor 1-alpha (tef-1a) and the largest and second largest subunit of the RNA polymerase (rpb1, rpb2), O. ovatospora was clustered together with O. khokpasiensis to form a clade separate from other species of Ophiocordyceps. Morphologically, O. ovatospora is characterized by producing pseudo-immersed perithecia, filiform asci and ascospores, monophialidic phialides, and oval conidia. Ophiocordyceps ovatospora has smaller perithecia than those of O. khokpasiensis but its asci are larger.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Araújo, J.P.M., Evans, H.C., Geiser, D.M., Mackay, W.P. & Hughes, D.P. (2015) Unravelling the diversity behind the <em>Ophiocordyceps unilateralis</em> (Ophiocordycipitaceae) complex: three new species of zombie-ant fungi from the Brazilian Amazon. <em>Phytotaxa</em> 220: 224–238. https://doi.org/10.11646/phytotaxa.220.3.2</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Araújo, J.P.M., Evans, H.C., Kepler, R.M. & Hughes, D.P. (2018) Zombie-ant fungi across continents: 15 new species and new combinations within <em>Ophiocordyceps</em>. I. <em>Myrmecophilous</em> hirsutelloid species. <em>Studies in Mycology</em> 90: 119–160. https://doi.org/10.1016/j.simyco.2017.12.002</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Bischoff, J.F., Rehner, S.A. & Humber, R.A. (2006) <em>Metarhizium frigidum</em> sp. nov.: a cryptic species of<em> M. anisopliae</em> and a member of the <em>M. flavoviride</em> complex. <em>Mycologia</em> 98: 737–745. https://doi.org/10.3852/mycologia.98.5.737</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Blackwell, M. & Gilbertson, R.L. (1981) <em>Cordycepioideus octosporus</em>, a termite suspected pathogen from Jalisco, Mexico. <em>Mycologia</em> 73: 358–362. https://doi.org/10.2307/3759659</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Blackwell, M. & Gilbertson, R.L. (1984) New information on <em>Cordycepioideus bisporus</em> and <em>Cordycepioideus octosporus</em>. <em>Mycologia </em>76: 763–765. https://doi.org/10.2307/3793239</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Buckley, R.E. (2005) A <em>Beauveria</em> phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to <em>Cordyceps</em> teleomorphs. <em>Mycologia</em> 97: 84–98. https://doi.org/10.1080/15572536.2006.11832842</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Castlebury, L.A., Rossman, A.Y., Sung, G.H., Hyten, A.S. & Spatafora, J.W. (2004) Multigene phylogeny reveals new lineage for <em>Stachybotrys chartarum</em>, the indoor air fungus. <em>Mycological Research</em> 108: 864–872. https://doi.org/10.1017/S0953756204000607</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Eggleton, P. (2000) <em>Termites: evolutions, sociality symbioses, ecology.</em> Kluwer Academic Publishers, Dordrecht. pp. 25–51. </span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Evans, H.C., Elliot, S.L., Hughes, D.P. & Corrie, M. (2011) Hidden diversity behind the zombie-ant fungus <em>Ophiocordyceps unilateralis</em>: four new species described from carpenter ants in Minas Gerais, Brazil. <em>PLoS One</em> 6: e17024. https://doi.org/10.1371/journal.pone.0017024</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Khonsanit, A., Luangsa-ard, J.J., Thanakitpipattana, D., Kobmoo, N. & Piasai1, O. (2018) Cryptic species within <em>Ophiocordyceps myrmecophila</em> complex on formicine ants from Thailand. <em>Mycological Progress</em> 18: 147–161. https://doi.org/10.1007/s11557-018-1412-7</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kirk, P.M., Cannon, P.F., Minter, D.W. & Stalpers, J.A. (2008) <em>Ainsworth & Bisby’s dictionary of the fungi</em>, 10th edn. CABI International, Wallingford. 771 pp.</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kobayasi, Y. (1941) The genus <em>Cordyceps</em> and its allies. <em>Science Reports of the Tokyo Bunrika Daigaku </em>84: 53–260.</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kobmoo, N., Mongkolsamrit, S., Tasathai, K., Thanakitpipattana, D. & Luangsa-ard, J.J. (2012) Molecular phylogenies reveal host-specific divergence of <em>Ophiocordyceps unilateralis</em> sensu lato following its host ants. <em>Molecular Ecology</em> 21: 3022–3031. https://doi.org/10.1111/j.1365-294X.2012.05574.x</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kobmoo, N., Mongkolsamrit, S., Wutikhun, T., Tasanathai, K., Khonsanit, A., Thanakitpipattana, D. & Luangsa-Ard, T.T. (2015) New species of <em>Ophiocordyceps unilateralis</em>, an ubiquitous pathogen of ants from Thailand. <em>Fungal Biology</em> 119: 44–52. https://doi.org/10.1016/j.funbio.2014.10.008</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. & Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. <em>Bioinformatics</em> 23: 2947–2948. https://doi.org/10.1093/bioinformatics/btm404</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. <em>Molecular Biology and Evolution</em> 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, Z., Liang, Z.Q., Whalley, A.J.S., Yao, Y.J. & Liu, A.Y. (2001) <em>Cordyceps</em> <em>brittlebankisoides</em>, a new pathogen of grubs and its anamorph, <em>Metarhizium anisopliae</em> var.<em> majus</em>. <em>Journal of Invertebrate Pathology</em> 78: 178–182. https://doi.org/10.1006/jipa.2001.5039</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Luangsa-ard, J.J., Ridkaew, R., Tasanathai, K., Thanakitpipattana, D. & Hywel-Jones, N. (2010) <em>Ophiocordyceps halabalaensis</em>: a new species of <em>Ophiocordyceps </em>pathogenic to <em>Camponotus gigas</em> in Hala Bala Wildlife Sanctuary, Southern Thailand. <em>Fungal Biology</em> 115: 608–614. https://doi.org/10.1016/j.funbio.2011.03.002</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Luangsa-ard, J.J., Tasanathai, K., Thanakitpipattana, D., Khonsanit, A. & Stadler, M. (2018) Novel and interesting <em>Ophiocordyceps</em> spp. (Ophiocordycipitaceae, Hypocreales) with superficial perithecia from Thailand. <em>Studies in Mycology</em> 89: 125–142. https://doi.org/10.1016/j.simyco.2018.02.001</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ochiel, G.S., Evans, H.C. & Eilenberg, J. (1997) <em>Cordycepioideus</em>, a pathogen of termites in Kenya. <em>Mycologist</em> 11: 7–9. https://doi.org/10.1016/S0269-915X(97)80059-6</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Pearce, M. & Pearce, M. (1997) Termites: biology and pest management. <em>Ecological Entomology</em> 23: 354–354. https://doi.org/10.1046/j.1365-2311.1998.00139.x</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Petch, T. (1931) Notes on entomogenous fungi. <em>Transactions of the British</em> <em>Mycological Society</em> 16: 55–75. https://doi.org/10.1016/S0007-1536(31)80006-3</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rath, A.C. (2000) The use of entomopathogenic fungi for control of termites. <em>Biocontrol Science and Technology</em> 10: 563–581. https://doi.org/10.1080/095831500750016370.</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rehner, S.A. & Samuels, G.J. (1994) Taxonomy and phylogeny of <em>Gliocladium</em> analysed from nuclear large subunit ribosomal DNA sequences. <em>Mycological Research </em>98: 625–634. https://doi.org/10.1016/S0953-7562(09)80409-7</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models.<em> Bioinformatics</em> 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Spatafora, J.W., Quandt, C.A., Kepler, R.M., Sung, G.H., Shrestha, B., Hywel-Jones, N.L. & Luangsa-ard, J.J. (2015) New 1F1N species combinations in Ophiocordycipitaceae (Hypocreales). <em>IMA Fungus</em> 6: 357–362. https://doi.org/10.5598/imafungus.2015.06.02.07</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. <em>Bioinformatics </em>22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stifler, C.B. (1941) A new genus of Hypocreales. <em>Mycologia </em>33: 82–86.</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Sung, G.H., Hywel-Jones, N.L., Sung, J.M., Luangsa-ard, J.J., Shrestha, B. & Spatafora, J.W. (2007) Phylogenetic classification of <em>Cordyceps </em>and the clavicipitaceous fungi.<em> Studies in Mycology</em> 57: 5–59. https://doi.org/10.3114/sim.2007.57.01</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Swofford, D.L. (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts. https://doi.org/10.1111/j.0014-3820.2002.tb00191.x</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. <em>Molecular Biology and Evolutio</em>n 30: 2725–2729.</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tasanathai, K., Noisripoom, W., Chaitika, T., Khonsanit, A., Hasin, S. & Luangsa-ard, J. (2019) Phylogenetic and morphological classification of <em>Ophiocordyceps</em> species on termites from Thailand. <em>MycoKeys</em> 56: 101–129. https://doi.org/10.3897/mycokeys.56.37636</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several<em> Cryptococcus</em> species. <em>Journal of Bacteriology</em> 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990 </span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wang, Y.B., Yu, H., Dai, Y.D., Chen, Z.H., Zeng, W.B., Yuan, F. & Liang, Z.Q. (2015) <em>Polycephalomyces yunnanensis</em> (Hypocreales), a new species of <em>Polycephalomyces </em>parasitizing <em>Ophiocordyceps nutans</em> and stink bugs (hemipteran adults). <em>Phytotaxa</em> 208: 34–44. https://doi.org/10.11646/phytotaxa.208.1.3</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wang, Y.B., Wang, Y., Fan, Q., Duan, D.E., Zhang, G.D., Dai, R.Q., Dai, Y.D., Zeng, W.B., Chen, Z.H., Li, D.D., Tang, D.X., Xu, Z.H., Sun, T., Nguyen, T.T., Tran, N.L., Dao, V.M., Zhang, C.M., Huang, L.D., Liu, Y.J., Zhang, X.M., Yang, D.R., Sanjuan, T., Liu, X.Z., Yang, Z.L. & Yu, H. (2020) Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus <em>Paecilomyces hepiali</em>. <em>Fungal Diversity</em> 103: 1–46. https://doi.org/10.1007/S13225-020-00457-3</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>PCR Protocols</em> 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1</span></span></span></p>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Xu, Z.H., Tran, N.L., Wang, Y., Zhang, G.D., Dao, V.M., Nguyen, T.T., Wang, Y.B. & Yu, H. (2022) Phylogeny and morphology of <em>Ophiocordyceps puluongensis</em> sp. nov. (Ophiocordycipitaceae, Hypocreales), a new fungal pathogen on termites from Vietnam. <em>Journal of Invertebrate Pathology </em>192: 107771. </span></span></span><span style="font-size: small; font-family: 'Times New Roman', serif;">https://doi.org/10.1016/j.jip.2022.107771</span></p>