Abstract
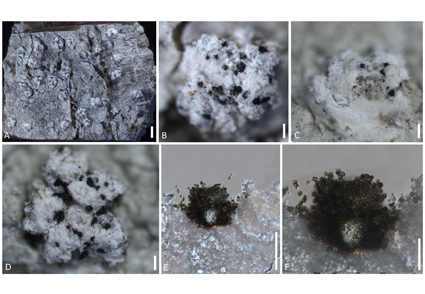
Sclerococcum is a species rich lichenicolous genus in Ascomycota. However, many species of Sclerococcum have been described based only on morphology. This study provides sequences for the large-subunit ribosomal RNA (LSU), internal transcribed spacer (ITS) and mitochondrial small-subunit ribosomal RNA (mtSSU) for the first time for Sclerococcum simplex, and the first geographical record of the genus from China. Phylogenetic analyses confirmed that S. simplex clustered within Sclerococcum sensu stricto while the genus was recovered as paraphyletic in Dactylosporaceae. Our asexual specimen of Sclerococcum simplex was collected on a corticolous Pertusaria thallus. The taxonomic affinity of Sclerococcum simplex is investigated based on phylogenetic and morphological evidence.
References
<p>Diederich, P. & van den Boom, P.P.G. (2013) Two new lichenicolous species of <em>Hainesia</em> (asexual Ascomycetes) growing on <em>Cladonia</em>. <em>Bulletin de la Societé des naturalistes luxembourgeois</em> 114: 59–64.</p>
<p>Diederich, P. & van den Boom, P.P.G. (2017) <em>Sclerococcum phaeophysciae</em> and <em>S. toensbergii</em>, two new lichenicolous asexual Ascomycetes, with a revised key to the species of <em>Sclerococcum</em>. <em>Bulletin de la Société des naturalistes luxembourgeois</em> 119: 71–78.</p>
<p>Diederich, P. (1990) New or interesting lichenicolous fungi 1. Species from Luxembourg. <em>Mycotaxon </em>37: 297–330.</p>
<p>Diederich, P. (2010) <em>Sclerococcum cladoniae</em>, a new lichenicolous hyphomycete on <em>Cladonia</em> from Luxembourg. <em>Bulletin de la Societé des naturalistes luxembourgeois </em>111: 57–59.</p>
<p>Diederich, P. (2015) Two new lichenicolous species of <em>Sclerococcum </em>(asexual Ascomycetes) growing on Graphidaceae. <em>Bulletin de la Société des naturalistes luxembourgeois</em> 117: 35–42.</p>
<p>Diederich, P., Ertz, D., Lawrey, J.D., Sikaroodi, M. & Untereiner, W.A. (2013) Molecular data place the hyphomycetous lichenicolous genus <em>Sclerococcum </em>close to <em>Dactylospora</em> (Eurotiomycetes) and <em>S. parmeliae</em> in <em>Cladophialophora </em>(Chaetothyriales). <em>Fungal Diversity</em> 58: 61–72. https://doi.org/10.1007/s13225-012-0179-4</p>
<p>Diederich, P. & van den Boom, P. (2017) <em>Sclerococcum phaeophysciae</em> and <em>S. toensbergii</em>, two new lichenicolous asexual Ascomycetes, with a revised key to the species of <em>Sclerococcum</em>. <em>Bulletin de la Société des naturalistes luxembourgeois</em> 119: 71–78.</p>
<p>Diederich, P., Lawrey, J.D. & Ertz, D. (2018) The 2018 classification and checklist of lichenicolous fungi, with 2000 non-lichenized, obligately lichenicolous taxa. <em>Bryologist </em>121: 340–425. https://doi.org/10.1639/0007-2745-121.3.340</p>
<p>Dissanayake, A.J., Bhunjun, C.S., Maharachchikumbura, S.S.N. & Liu, J.K. (2020) Applied aspects of methods to infer phylogenetic relationships amongst fungi. <em>Mycosphere </em>11 (1): 2652–2676. https://doi 10.5943/mycosphere/11/1/18</p>
<p>Dong, W., Hyde, K.D., Doilom, M., Yu, X.D., Bhat, D.J., Jeewon, R., Boonmee, S., Wang, G.N., Nalumpang, S. & Zhang, H. (2020) <em>Pseudobactrodesmium </em>(Dactylosporaceae, Eurotiomycetes, Fungi) a novel lignicolous genus. <em>Frontiers in Microbiology</em> 11: 1–13. https://doi.org/10.3389/fmicb.2020.00456</p>
<p>Ekanayaka, A.H., Jones, E.G., Hyde, K.D. & Zhao, Q. (2019) A stable phylogeny for Dactylosporaceae. <em>Cryptogamie Mycologie</em> 40 (3): 23–44. https://doi.org/10.5252/cryptogamie-mycologie2019v40a3</p>
<p>Etayo, J. & Diederich, P. (1995) Lichenicolous fungi from the western Pyrenees, France and Spain. I. Flechten Follmann. <em>Contributions to lichenology in honour of Gerhard Follmann </em>205–221.</p>
<p>Fries, E.M. (1819) <em>Novitiae florae suecicae</em> 5. Lund.</p>
<p>Fries, E.M. (1825) <em>Systema orbis vegetabilis</em> I. Lund.</p>
<p>GenBank (2021) Available from: https://www.ncbi.nlm.nih.gov/nuccore/ (accessed 6 April 2021)</p>
<p>Glez-Peña, D., Gómez-Blanco, D., Reboiro-Jato, M., Fdez-Riverola, F. & Posada, D. (2010) ALTER: program-oriented conversion of DNA and protein alignments. <em>Nucleic Acids Research</em> 38: 14–18. https://doi.org/10.1093/nar/gkq321</p>
<p>Hawksworth, D.L. (1979) The lichenicolous Hyphomycetes. Bulletin of the British Museum (Natural History). <em>Botany Series</em> 6: 183–300.</p>
<p>Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. <em>Bioinformatics</em> 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754</p>
<p>Index Fungorum (2021) Available from: http://www.indexfungorum.org/names/Names.asp (accessed 6 April 2021)</p>
<p>Jayasiri, S.C., Hyde, K.D., Ariyawansa, H.A., Bhat, J., Buyck, B., Cai, L., Dai, Y.C., Abd-Elsalam, K.A., Ertz, D., Hidayat, I., Jeewon, R., Jones, E.B.G., Bahkali, A.H., Karunarathna, S.C., Liu, J.K., Luangsa-ard, J.J., Lumbsch, H.T. Maharachchikumbura, S.S.N., McKenzie, E.H.C., Moncalvo, J.M., Ghobad-Nejhad, M., Nilsson, H., Pang, K.A., Pereira, O.L., Phillips, A.J.L., Raspé, O., Rollins, A.W., Romero, A.I., Etayo, J., Selçuk, F., Stephenson, S.L., Suetrong, S., Taylor, J.E., Tsui, C.K.M., Vizzini, A., Abdel-Wahab, M.A., Wen, T.C., Boonmee, S., Dai, D.Q., Daranagama, D.A., Dissanayake, A.J., Ekanayaka, A.H., Fryar, S.C., Hongsanan, S., Jayawardena, R.S., Li, W.J., Perera, R.H., Phookamsak, R., de Silva, N.I., Thambugala, K.M., Tian, Q., Wijayawardene, N.N., Zhao, R.L., Zhao, Q., Kang, J.C. & Promputtha, I. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. <em>Fungal Diversity </em>74 (1): 3–18. https://doi.org/10.1007/s13225-015-0351-8</p>
<p>Joshi, Y. (2021) Two new species of lichenicolous fungus <em>Sclerococcum</em> (Dactylosporaceae, Sclerococcales) from India. <em>Acta Botanica Hungarica</em> 63: 67–75. https://doi.org/10.1556/034.63.2021.1-2.5</p>
<p>Joshi, Y., Falswal, A., Tripathi, M., Upadhyay, S., Bisht, A., Chandra, K., Bajpai, R. & Upreti, D.K. (2016) One hundred and five species of lichenicolous biota from India: An updated checklist for the country. <em>Mycosphere</em> 7: 268–294. https://doi.org/10.5943/mycosphere/7/3/3</p>
<p>Katoh, K. & Standley, K. (2013) MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. <em>Molecular Biology. Evolution </em>30: 772–780. https://doi.org/10.1093/molbev/mst010</p>
<p>Lawrey, J.D., Binder, M., Diederich, P., Molina, M.C., Sikaroodi, M. & Ertz, D. (2007) Phylogenetic diversity of lichen-associated homobasidiomycetes. <em>Molecular Phylogenetics and Evolution</em> 44 (2): 778–789. https://doi.org/10.1016/j.ympev.2006.12.023</p>
<p>Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees.<em> In: SC10 Workshop on Gateway Computing Environments (GCE10).</em> https://doi.org/10.1109/GCE.2010.5676129</p>
<p>Nylander, J.A.A. (2004) MrAIC. pl. Program distributed by the author. Sweden: Evolutionary Biology Centre, Uppsala University.</p>
<p>Olariaga, I., Teres, J., Martín, J., Prieto, M. & Baral, H.O. (2019) <em>Pseudosclerococcum golindoi</em> gen. et sp. nov., a new taxon with apothecial ascomata and a <em>Chalara</em>-like anamorph within the Sclerococcales (Eurotiomycetes). <em>Mycological Progress </em>18: 895–905. https://doi.org/10.1007/s11557-019-01500-7</p>
<p>Pang, K.L., Guo, S.Y., Alias, S.A., Hafellner, J. & Jones, E.B.G. (2014) A new species of marine <em>Dactylospora</em> and its phylogenetic affinities within the Eurotiomycetes, Ascomycota. <em>Botanica Marina</em> 57: 315–321. https://doi.org/10.1515/bot-2014-0025</p>
<p>Pino-Bodas, R., Zhurbenko, M.P. & Stenroos, S. (2017) Phylogenetic placement within Lecanoromycetes of lichenicolous fungi associated with <em>Cladonia</em> and some other genera. <em>Persoonia</em> 39: 91–117.</p>
<p>Rambaut, A. (2012) FigTree version 1.4.0. Available at http://tree.bio.ed.ac.uk/software/figtree/ (accessed 1 April 2021)</p>
<p>Réblová, M., Untereiner, W.A., Štěpánek, V. & Gams, W. (2017) Disentangling <em>Phialophora</em> section Catenulatae: disposition of taxa with pigmented conidiophores and recognition of a new subclass, Sclerococcomycetidae (Eurotiomycetes). <em>Mycological Progress</em> 16: 27–46. https://doi.org/10.1007/s11557-016-1248-y</p>
<p>Senanayake, I.C., Rathnayake, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. <em>Mycosphere</em> 11 (1): 2678–2754. https://doi 10.5943/mycosphere/11/1/20</p>
<p>Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. <em>Bioinformatics</em> 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</p>
<p>van den Boom, P.P.G., Breuss, O., Spier, L. & Brand, A. (1996) Beitrag zur Flechtenflora Kärntens. Ergebnisse der Feldtagung der bryologischen und lichenologischen Arbeitsgruppe der KNNV in Weissbriach. <em>Linzer biologische Beiträge</em> 28: 619–654.</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus </em>species. <em>Journal of Bacteriology</em> 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990</p>
<p>Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T., Tedersoo, L., Haelewaters, D., Rajeshkumar, K.C., Zhao, R.L., Aptroot, A., Leontyev, D.V., Saxena, R.K., Tokarev, Y.S., Dai, D.Q, Letcher, P.M, Stephenson, S.L., Ertz, D., Lumbsch, H.T., Kukwa, M., Issi, I.V., Madrid, H., Phillips, A.J.L, Selbmann, L, Pfliegler, W.P, Horváth, E, Bensch, K., Kirk, P., Kolaříková, Z., Raja, H.A., Radek, R, Papp, V., Dima, B., Ma, J., Malosso, E., Takamatsu, S., Rambold, G., Gannibal, P.B., Triebel, D., Gautam, A.K., Avasthi, S., Suetrong, S., Timdal, E., Fryar, S.C., Delgado, G., Réblová, M., Doilom, M., Dolatabadi, S., Pawłowska, J., Humber, R.A., Kodsueb, R., Sánchez-Castrov, I., Goto, B.T., Silva, D.K.A., de Souza, F.A., Oehl, F., da Silva, G.A., Silva, I.R., Błaszkowski, J., Jobim, K., Maia, L.C., Barbosa, F.R., Fiuza, P.O., Divakar, P.K., Shenoy, B.D., Castañeda-Ruiz, R.F., Somrithipol, S., Karunarathna, S.C., Tibpromma, S., Mortimer,.PE., Wanasinghe, D.N., Phookamsak, R., Xu, J., Wang, Y., Fenghua, T., Alvarado, P., Li, D.W., Kušan, I., Matočec, N., Maharachchikumbura, S.S.N., Papizadeh, M., Heredia, G., Wartchow, F., Bakhshi, M., Boehm, E., Youssef, N., Hustad, V.P., Lawrey, J.D., Santiago, A.L.C.M., Bezerra, J.D.P., Souza-Motta, C.M., Firmino, A.L., Tian, Q., Houbraken, J., Hongsanan, S., Tanaka, K., Dissanayake, A.J., Monteiro, J.S., Grossart, H.P., Suija, A., Weerakoon, G., Etayo, J., Tsurykau, A., Kuhnert, E., Vázquez, V., Mungai, P., Damm, U., Li, Q.R., Zhang, H., Boonmee, S., Lu, Y.Z., Becerra AG., Kendrick, B., Brearley FQ., Motiejűnaitë, J., Sharma, B., Khare, R., Gaikwad, S., Wijesundara, D.S.A., Tang, L.Z., He, M.Q., Flakus, A., Rodriguez-Flakus, P., Zhurbenko, M.P., McKenzie, E.H.C., Stadler, M., Bhat, D.J., Kui-Liu, J., Raza, M., Jeewon, R., Nassonova, E.S., Prieto, M., Jayalal, R.G.U., Erdoðdu, M., Yurkov, A., Schnittler, M., Shchepin, O.N., Novozhilov, Y.K., Silva-Filho, AGS., Gentekaki, E., Liu, P., Cavender, J.C., Kang, Y., Mohammad, S., Zhang, L.F., Xu, R.F., Li, Y.M., Dayarathne, M.C., Ekanayaka, A.H., Wen, T.C., Deng, C.Y., Lateef, A.A., Pereira, O.L., Navathe, S., Hawksworth, D.L., Fan, X.L., Dissanayake, L.S., Kuhnert, E., Grossart, H.P. & Thines, M. (2020) Outline of fungi and fungus-like taxa. <em>Mycosphere</em> 11: 1060–1456. https://doi.org/10.5943/mycosphere/11/1/8</p>
<p>Yu, X.D., Dong, W., Bhat, D.J., Boonmee, S., Zhang, D.I. & Zhang, H. (2018) <em>Cylindroconidiis aquaticus</em> gen. et sp. nov., a new lineage of aquatic hyphomycetes in Sclerococcaceae (Eurotiomycetes). <em>Phytotaxa </em>372: 79–87. https://doi.org/10.11646/phytotaxa.372.1.6</p>
<p>Zhurbenko, M.P. (2004) Lichenicolous and some interesting lichenized fungi from the Northern Ural, Komi Republic of Russia. <em>Herzogia </em>17: 77–86.</p>
<p>Zoller, S., Scheidegger, C. & Sperisen, C. (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. <em>Lichenologist </em>31: 511–516. https://doi.org/10.1006/lich.1999.0220</p>