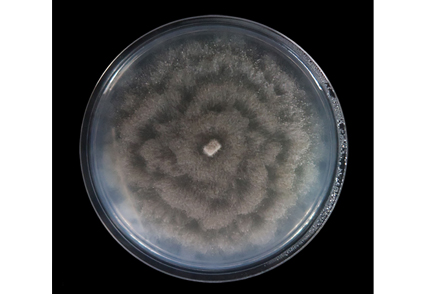
Abstract
Coniella has a wide distribution as plant pathogens, saprobes and endophytes. Some Coniella species infect leaves, fruits, stems and roots of plants. A coniella-like species was isolated from symptomatic leaves of Castanea mollissima (Fagaceae) from orchard in Tai’an City, Shandong Province, China. Based on multi-locus phylogenetic analyses of ITS, LSU, RPB2 and TEF1-α sequence data and morphology, Coniella castanea sp. nov. is introduced. A detailed description and illustration has been provided and new specieswas compared with related taxa.
References
<p>Alvarez, L.V., Groenewald, J.Z. & Crous, P.W. (2016) Revising the Schizoparmaceae: <em>Coniella</em> and its synonyms <em>Pilidiella</em> and <em>Schizoparme</em>. <em>Studies in Mycology </em>85: 1–34. https://doi.org/10.1016/j.simyco.2016.09.001</p>
<p>Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. <em>Mycologia</em> 91 (3): 553–556. https://doi.org/10.1080/00275514.1999.12061051</p>
<p>Chethana, K.W.T., Zhou, Y., Zhang, W., Liu, M., Xing, Q.K., Li, X.H., Yan, J.Y., Chethana, K.W.T. & Hyde, K.D. (2017) <em>Coniella vitis </em>sp. nov. Is the Common Pathogen of White Rot in Chinese Vineyards. <em>Plant Disease </em>101: 2123–2136. https://doi.org/10.1094/PDIS-12-16-1741-RE</p>
<p>Fröhlich, J. & Hyde, K.D. (2000) Palm microfungi. <em>Fungal Diversity Research Series </em>3: 1–375.</p>
<p>Gao, Y.H., Sun, W., Su, Y.Y. & Cai, L. (2013) Three new species of <em>Phomopsis</em> in Gutianshan Nature Reserve in China. <em>Mycological Progress </em>13: 111–121. https://doi.org/10.1007/s11557-013-0898-2</p>
<p>Guo, L.D., Hyde, K.D. & Liew, E.C.Y. (2000) Identification of endophytic fungi from <em>Livistona chinensis</em> based on morphology and rDNA sequences. <em>New Phytologist</em> 147 (3): 617–630. https://doi.org/10.1046/j.1469-8137.2000.00716.x</p>
<p>Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogeny trees. <em>Bioinformatics</em> 17 (17): 754–755. https://doi.org/10.1093/bioinformatics/17.8.754</p>
<p>Jayawardena, R.S., Purahong, W., Zhang, W., Wubet, T., Li, X., Liu, M., Zhao, W., Hyde, K.D., Liu, J. & Yan, J. (2018) Biodiversity of fungi on <em>Vitis vinifera</em> L. revealed by traditional and high-resolution culture-independent approaches. <em>Fungal Diversity </em>90: 1–84. https://doi.org/10.1007/s13225-018-0398-4</p>
<p>Jiang, N., Liang, L.Y. & Tian, C.M. (2020) <em>Gnomoniopsis chinensis</em> (Gnomoniaceae, Diaporthales), a new fungus causing canker of Chinese chestnut in Hebei Province, China. <em>MycoKeys </em>67: 19–32. https://doi.org/10.3897/mycokeys.67.51133</p>
<p>Jiang, N., Voglmayr, H., Piao, C.G. & Li, Y. (2021a) Two new species of <em>Diaporthe</em> (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. <em>MycoKeys</em> 85: 31–56. https://doi.org/10.3897/mycokeys.85.73107</p>
<p>Jiang, N., Voglmayr, H., Bian, D.R., Piao, C.G., Wang, S.K. & Li, Y. (2021b) Morphology and Phylogeny of <em>Gnomoniopsis</em> (Gnomoniaceae, Diaporthales) from Fagaceae Leaves in China. <em>Journal of Fungi</em> 7: 792. https://doi.org/10.3390/jof7100792</p>
<p>Jiang, S.X., Liu, C.Z., Wang, Q.H., Jia, N., Li, C. & Ma, H.B. (2011) A New Chestnut Disease—Brown Margin Leaf Blight and the Pathogen Identification. <em>Scientia Silvae Sinicae </em>47 (5): 177–180. https://doi.org/10.1007/s11676-011-0141-4</p>
<p>Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. <em>Briefings in Bioinformatics</em> 20: 1160–1166. https://doi.org/10.1093/bib/bbx108</p>
<p>Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. <em>Molecular Biology and Evolution</em> 33 (7): 1870–1874. https://doi.org/10.1093/molbev/msw054</p>
<p>Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic Relationships among Ascomycetes: evidence from an RNA polymerse II subunit. <em>Molecular Biology and Evolution</em> 16 (12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092</p>
<p>Miller, M.A., Pfeiffer, W. & Schwartz, T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources.<em> In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond.</em> Chicago, Illinois, USA: Association for Computing Machinery, pp. Article 39, 1–8. https://doi.org/10.1145/2335755.2335836</p>
<p>Muthumary, J. & Vanaja, R. (1986) Development of conidiomata in <em>Coniella fragariae.</em> <em>Transactions of the British Mycological Society</em> 87: 109–113. https://doi.org/10.1016/S0007-1536(86)80009-2</p>
<p>Nag Raj, T.R. (1993) <em>Coelomycetous anamorphs with appendage-bearing conidia</em>. Mycologue Publications, Waterloo, Canada.</p>
<p>Nylander, J.A.A. (2004) <em>MrModelTest V2</em>, <em>Program Distributed by the Author</em>. Evolutionary Biology Centre, Uppsala University.</p>
<p>Pollastro, S., Dongiovanni, C., Gerin, D., Pollastro, P., Fumarola, G., De Miccolis Angelini, R.M. & Faretra, F. (2016) First Report of <em>Coniella granati</em> as a Causal Agent of Pomegranate Crown Rot in Southern Italy. <em>Plant Disease </em>100 (7): 1498. https://doi.org/10.1094/PDIS-11-15-1320-PDN</p>
<p>Rehner, S.A. & Samuels, G.J. (1994) Taxonomy and phylogeny of <em>Gliocladium</em> analysed from nuclear large subunit ribosomal DNA sequences. <em>Mycological Research</em> 98 (6): 625–634. https://doi.org/10.1016/S0953-7562(09)80409-7</p>
<p>Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. <em>Bioinformatics</em> 19 (12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180</p>
<p>Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. <em>Systematic Biology </em>61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Rossman, A.Y., Farr, D.F. & Castlebury, L.A. (2007) A review of the phylogeny and biology of the Diaporthales. <em>Mycoscience </em>48: 135–144. https://doi.org/10.1007/s10267-007-0347-7</p>
<p>Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J., Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. <em>Mycosphere</em> 11 (1): 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20</p>
<p>Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. <em>Bioinformatics </em>30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</p>
<p>Sun, W.X., Huang, S.T., Xia, J.W., Zhang, X.G. & Li, Z. (2021) Morphological and molecular identification of <em>Diaporthe</em> species in south-western China, with description of eight new species. <em>MycoKeys </em>77: 65–95. https://doi.org/10.3897/mycokeys.77.59852</p>
<p>Sutton, B.C. (1980) <em>The Coelomycetes</em>. <em>Fungi imperfecti with pycnidia, acervuli and stromata</em>. Commonwealth Mycological Institute, Kew, UK.</p>
<p>Tennakoon, D.S., Kuo, C.H., Maharachchikumbura, S.S.N., Thambugala, K.M., Gentekaki, E., Phillips, A.J.L., Bhat, D.J., Wanasinghe, D.N., de Silva, N.I., Promputtha, I. & Hyde, K.D. (2021) Taxonomic and phylogenetic contributions to <em>Celtis formosana</em>, <em>Ficus ampelas</em>, <em>F. septica</em>, <em>Macaranga tanarius</em> and <em>Morus australis</em> leaf litter inhabiting microfungi. <em>Fungal Diversity </em>108: 1–215. https://doi.org/10.1007/s13225-021-00474-w</p>
<p>Van Niekerk, J.M., Groenewald, J.Z., Verkley, G.J., Fourie, P.H., Wingfield, M.J. & Crous, P.W. (2004) Systematic reappraisal of <em>Coniella</em> and <em>Pilidiella</em>, with specific reference to species occurring on Eucalyptus and Vitis in South Africa. <em>Mycological Research </em>108: 283–303 https://doi.org/10.1017/s0953756204009268</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus</em> species. <em>Journal of Bacteriology</em> 172 (8): 4238–4246. https://doi.org/10.1128/JB.172.8.4238-4246.1990</p>
<p>Von Arx, J.A. (1973) Centraalbureau voor Schimmelcultures Baarn and Delft. Progress Report 1972. <em>Verhandelingen der Koninklijke Nederlandsche Akademie van Wetenschappen, Afdeling Natuurkunde</em> 61: 59–81.</p>
<p>Von Arx, J.A. (1981) <em>The genera of fungi sporulating in pure culture</em>, 3rd edn. J Cramer, Vaduz.</p>
<p>Von Höhnel, F. (1918) Dritte vorlaufige Mitteilung mycologischer Ergebnisse (Nr. 201–304). <em>Berichte der Deutschen Botanischen Gesellschaft</em> 36: 309–317.</p>
<p>White, T.J., Bruns, T.D., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>In:</em> Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) <em>PCR protocols, a guide to methods and applications.</em> Academic Press, San Diego, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</p>
<p>Zhuang, W.Y., Guo, L., Guo, S.Y., Guo, Y.L., Mao, X.L., Sun, S.X., Wei, S.X., Wen, H.A., Yu, Z.H. & Zhang, X.Q. (2001) <em>Higher fungi of tropical china</em>. Mycotaxon Ltd.Ithaca, New York, pp. 1–485.</p>
<p>Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. <em>Mycologia</em> 91 (3): 553–556. https://doi.org/10.1080/00275514.1999.12061051</p>
<p>Chethana, K.W.T., Zhou, Y., Zhang, W., Liu, M., Xing, Q.K., Li, X.H., Yan, J.Y., Chethana, K.W.T. & Hyde, K.D. (2017) <em>Coniella vitis </em>sp. nov. Is the Common Pathogen of White Rot in Chinese Vineyards. <em>Plant Disease </em>101: 2123–2136. https://doi.org/10.1094/PDIS-12-16-1741-RE</p>
<p>Fröhlich, J. & Hyde, K.D. (2000) Palm microfungi. <em>Fungal Diversity Research Series </em>3: 1–375.</p>
<p>Gao, Y.H., Sun, W., Su, Y.Y. & Cai, L. (2013) Three new species of <em>Phomopsis</em> in Gutianshan Nature Reserve in China. <em>Mycological Progress </em>13: 111–121. https://doi.org/10.1007/s11557-013-0898-2</p>
<p>Guo, L.D., Hyde, K.D. & Liew, E.C.Y. (2000) Identification of endophytic fungi from <em>Livistona chinensis</em> based on morphology and rDNA sequences. <em>New Phytologist</em> 147 (3): 617–630. https://doi.org/10.1046/j.1469-8137.2000.00716.x</p>
<p>Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogeny trees. <em>Bioinformatics</em> 17 (17): 754–755. https://doi.org/10.1093/bioinformatics/17.8.754</p>
<p>Jayawardena, R.S., Purahong, W., Zhang, W., Wubet, T., Li, X., Liu, M., Zhao, W., Hyde, K.D., Liu, J. & Yan, J. (2018) Biodiversity of fungi on <em>Vitis vinifera</em> L. revealed by traditional and high-resolution culture-independent approaches. <em>Fungal Diversity </em>90: 1–84. https://doi.org/10.1007/s13225-018-0398-4</p>
<p>Jiang, N., Liang, L.Y. & Tian, C.M. (2020) <em>Gnomoniopsis chinensis</em> (Gnomoniaceae, Diaporthales), a new fungus causing canker of Chinese chestnut in Hebei Province, China. <em>MycoKeys </em>67: 19–32. https://doi.org/10.3897/mycokeys.67.51133</p>
<p>Jiang, N., Voglmayr, H., Piao, C.G. & Li, Y. (2021a) Two new species of <em>Diaporthe</em> (Diaporthaceae, Diaporthales) associated with tree cankers in the Netherlands. <em>MycoKeys</em> 85: 31–56. https://doi.org/10.3897/mycokeys.85.73107</p>
<p>Jiang, N., Voglmayr, H., Bian, D.R., Piao, C.G., Wang, S.K. & Li, Y. (2021b) Morphology and Phylogeny of <em>Gnomoniopsis</em> (Gnomoniaceae, Diaporthales) from Fagaceae Leaves in China. <em>Journal of Fungi</em> 7: 792. https://doi.org/10.3390/jof7100792</p>
<p>Jiang, S.X., Liu, C.Z., Wang, Q.H., Jia, N., Li, C. & Ma, H.B. (2011) A New Chestnut Disease—Brown Margin Leaf Blight and the Pathogen Identification. <em>Scientia Silvae Sinicae </em>47 (5): 177–180. https://doi.org/10.1007/s11676-011-0141-4</p>
<p>Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. <em>Briefings in Bioinformatics</em> 20: 1160–1166. https://doi.org/10.1093/bib/bbx108</p>
<p>Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. <em>Molecular Biology and Evolution</em> 33 (7): 1870–1874. https://doi.org/10.1093/molbev/msw054</p>
<p>Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic Relationships among Ascomycetes: evidence from an RNA polymerse II subunit. <em>Molecular Biology and Evolution</em> 16 (12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092</p>
<p>Miller, M.A., Pfeiffer, W. & Schwartz, T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources.<em> In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond.</em> Chicago, Illinois, USA: Association for Computing Machinery, pp. Article 39, 1–8. https://doi.org/10.1145/2335755.2335836</p>
<p>Muthumary, J. & Vanaja, R. (1986) Development of conidiomata in <em>Coniella fragariae.</em> <em>Transactions of the British Mycological Society</em> 87: 109–113. https://doi.org/10.1016/S0007-1536(86)80009-2</p>
<p>Nag Raj, T.R. (1993) <em>Coelomycetous anamorphs with appendage-bearing conidia</em>. Mycologue Publications, Waterloo, Canada.</p>
<p>Nylander, J.A.A. (2004) <em>MrModelTest V2</em>, <em>Program Distributed by the Author</em>. Evolutionary Biology Centre, Uppsala University.</p>
<p>Pollastro, S., Dongiovanni, C., Gerin, D., Pollastro, P., Fumarola, G., De Miccolis Angelini, R.M. & Faretra, F. (2016) First Report of <em>Coniella granati</em> as a Causal Agent of Pomegranate Crown Rot in Southern Italy. <em>Plant Disease </em>100 (7): 1498. https://doi.org/10.1094/PDIS-11-15-1320-PDN</p>
<p>Rehner, S.A. & Samuels, G.J. (1994) Taxonomy and phylogeny of <em>Gliocladium</em> analysed from nuclear large subunit ribosomal DNA sequences. <em>Mycological Research</em> 98 (6): 625–634. https://doi.org/10.1016/S0953-7562(09)80409-7</p>
<p>Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. <em>Bioinformatics</em> 19 (12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180</p>
<p>Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. <em>Systematic Biology </em>61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Rossman, A.Y., Farr, D.F. & Castlebury, L.A. (2007) A review of the phylogeny and biology of the Diaporthales. <em>Mycoscience </em>48: 135–144. https://doi.org/10.1007/s10267-007-0347-7</p>
<p>Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J., Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. <em>Mycosphere</em> 11 (1): 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20</p>
<p>Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. <em>Bioinformatics </em>30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</p>
<p>Sun, W.X., Huang, S.T., Xia, J.W., Zhang, X.G. & Li, Z. (2021) Morphological and molecular identification of <em>Diaporthe</em> species in south-western China, with description of eight new species. <em>MycoKeys </em>77: 65–95. https://doi.org/10.3897/mycokeys.77.59852</p>
<p>Sutton, B.C. (1980) <em>The Coelomycetes</em>. <em>Fungi imperfecti with pycnidia, acervuli and stromata</em>. Commonwealth Mycological Institute, Kew, UK.</p>
<p>Tennakoon, D.S., Kuo, C.H., Maharachchikumbura, S.S.N., Thambugala, K.M., Gentekaki, E., Phillips, A.J.L., Bhat, D.J., Wanasinghe, D.N., de Silva, N.I., Promputtha, I. & Hyde, K.D. (2021) Taxonomic and phylogenetic contributions to <em>Celtis formosana</em>, <em>Ficus ampelas</em>, <em>F. septica</em>, <em>Macaranga tanarius</em> and <em>Morus australis</em> leaf litter inhabiting microfungi. <em>Fungal Diversity </em>108: 1–215. https://doi.org/10.1007/s13225-021-00474-w</p>
<p>Van Niekerk, J.M., Groenewald, J.Z., Verkley, G.J., Fourie, P.H., Wingfield, M.J. & Crous, P.W. (2004) Systematic reappraisal of <em>Coniella</em> and <em>Pilidiella</em>, with specific reference to species occurring on Eucalyptus and Vitis in South Africa. <em>Mycological Research </em>108: 283–303 https://doi.org/10.1017/s0953756204009268</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus</em> species. <em>Journal of Bacteriology</em> 172 (8): 4238–4246. https://doi.org/10.1128/JB.172.8.4238-4246.1990</p>
<p>Von Arx, J.A. (1973) Centraalbureau voor Schimmelcultures Baarn and Delft. Progress Report 1972. <em>Verhandelingen der Koninklijke Nederlandsche Akademie van Wetenschappen, Afdeling Natuurkunde</em> 61: 59–81.</p>
<p>Von Arx, J.A. (1981) <em>The genera of fungi sporulating in pure culture</em>, 3rd edn. J Cramer, Vaduz.</p>
<p>Von Höhnel, F. (1918) Dritte vorlaufige Mitteilung mycologischer Ergebnisse (Nr. 201–304). <em>Berichte der Deutschen Botanischen Gesellschaft</em> 36: 309–317.</p>
<p>White, T.J., Bruns, T.D., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>In:</em> Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) <em>PCR protocols, a guide to methods and applications.</em> Academic Press, San Diego, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</p>
<p>Zhuang, W.Y., Guo, L., Guo, S.Y., Guo, Y.L., Mao, X.L., Sun, S.X., Wei, S.X., Wen, H.A., Yu, Z.H. & Zhang, X.Q. (2001) <em>Higher fungi of tropical china</em>. Mycotaxon Ltd.Ithaca, New York, pp. 1–485.</p>