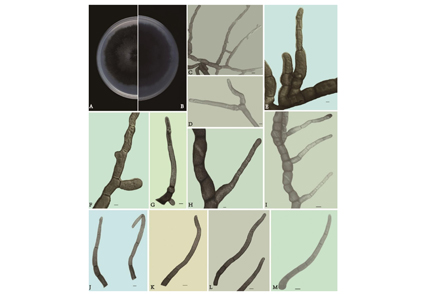
Abstract
The genus Phaeomycocentrospora (Dothidotthiaceae) is a fungal pathogen causing plant leaf spots. A new species, Phaeomycocentrospora xinjangensis sp. nov. is introduced from Xinjiang Uygur Autonomous Region, Northwest China. Multi-gene phylogenetic analyses based on a concatenated ITS, LSU, and ACT sequence dataset were used to confirm the phylogenetic position of the new species. Phaeomycocentrospora xinjangensis can easily be distinguished from the remaining species based on multi-gene phylogenetic analyses coupled with morphological characters. Morphologically, P. xinjangensis differs from other species in the genus by the presence of dark hyphae, dark brown conidiophores, few branched conidiogenous cells. Phylogenetically, our three strains were clustered together and formed a separate subclade with high support values. Moreover, we provided a description, illustrations, and phylogenetic tree for the new species.
References
<p>Carbone, I., Anderson, J.B. & Kohn, L.M. (1999) Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. <em>Evolution</em> 53: 11–21. https://10.1111/j.15585646.1999.tb05329.x</p>
<p>Crous, P.W., Braun, U., Hunter, G.C., Wingfield, M.J., Verkley, G.J.M., Shin, H.D., Nakashima, C. & Groenewald, J.Z. (2013) Phylogenetic lineages in <em>Pseudocercospora</em>.<em> Studies in Mycology</em> 75: 37–114. https://doi.org/10.3114/sim0005</p>
<p>Drummond, A. & Rambaut, A. (2007) BEAST: Bayesian evolutionary analysis by sampling trees. <em>BMC evolutionary biology</em> 7: 214–221. https://doi.org/10.1186/1471-2148-7-214</p>
<p>Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H.C., Sarma, V.V. & Xie, N. (2020) Refined families of dothideomycetes: dothideomycetidae and pleosporomycetidae. <em>Mycosphere</em> 11: 1553–2107. https://doi.org/10.5943/mycosphere/11/1/13</p>
<p>Katoh, K. & Standley, D.M. (2013) MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. <em>Molecular Biology and Evolution</em> 30: 772–780. https://doi.org/10.1093/molbev/mst010</p>
<p>Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. <em>Nature Methods</em> 14: 587–589. https://doi.org/10.1038/nmeth.4285</p>
<p>Li, X., Zhang, Z.Y., Chen, W.H., Liang, J.D., Huang, J.Z., Han, Y.F. & Liang, Z.Q. (2022) A new species of <em>Arthrographis</em> (Eremomycetaceae, Dothideomycetes), from the soil in Guizhou, China. <em>Phytotaxa</em> 538: 175–181. https://doi.org/10.11646/phytotaxa.538.3.1</p>
<p>Minh, Q., Nguyen, M. & von Haeseler, A.A. (2013) Ultrafast approximation for phylogenetic bootstrap. <em>Molecular Biology and Evolution</em> 30: 1188–1195. https://doi.org/10.1093/molbev/mst024</p>
<p>Marin-Felix, Y., Groenewald, J.Z., Cai, L., Chen, Q. & Crous, P.W. (2017) Genera of phytopathogenic fungi: gophy 1. <em>Studies in Mycology</em> 86: 99–216. https://doi.org/10.1016/j.simyco.2017.04.002</p>
<p>Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. <em>Molecular Biology and Evolution</em> 32: 268–274. https://doi.org/10.1093/molbev/msu300</p>
<p>Posada, D. & Crandall, K.A. (1998) Modeltest: testing the model of DNA substitution. <em>Bioinformatics</em>,14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817</p>
<p>Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. <em>Systematic Biology</em> 61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Rehner, S.A. & Samuels, G.J. (1994) Taxonomy and phylogeny of <em>Gliocladium</em> analysed from nuclear large subunit ribosomal DNA sequences. <em>Mycological Research</em> 98: 625–634. https://doi.org/10.1016/S0953-7562(09)80409-7</p>
<p>Senwanna, C., Wanasinghe, D.N., Bulgakov, T.S., Wang, Y., Bhat, D.J., Tang, A.M.C. & Phookamsak, R. (2019) Towards a natural classification of <em>Dothidotthia</em> and <em>Thyrostroma</em> in Dothidotthiaceae (Pleosporineae, Pleosporales). <em>Mycosphere</em> 10: 701–738. https://doi.org/10.5943/mycosphere/10/1/15</p>
<p>Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. <em>Molecular Biology and Evolution</em> 30: 2725–2729. https://doi.org/10.1093/molbev/mst197</p>
<p>Tullio, V., Banche, G., Allizond,V., Roana, J., Mandras, N., Scalas, D., Panzone, M., Cervetti, O., Valle, S., Carlone, N. & Cuffini, A.M. (2010) Non-dermatophyte moulds as skin and nail foot mycosis agents: <em>Phoma herbarum</em>, <em>Chaetomium globosum</em> and <em>Microascus cinereus</em>. <em>Fungal Biology</em> 114: 345–349. https://doi.org/10.1016/j.funbio.2010.02.003</p>
<p>Vaidya, G., Lohman, D.J. & Meier, R. (2011) Sequence Matrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. <em>Cladistics</em> 27: 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus</em> species. <em>Journal of Bacteriology</em> 172: 4238–4246. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC213247/pdf/jbacter00122-0118]</p>
<p>White, T.J., Bruns, T., Lee, S.J.W.T. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>PCR Protocols: a guide to methods and amplications</em> 18: 315–322. [https://msafungi.org/wp-content/uploads/2019/03/February-2013-Inoculum]</p>
<p>Zhang, Z.Y., Han, Y.F., Chen, W.H. & Liang, Z.Q. (2019) <em>Gongronella sichuanensis </em>(Cunninghamellaceae, Mucorales), a new species isolated from soil in China. <em>Phytotaxa</em> 416: 167–174. https://doi.org/10.11646/Phytotaxa.416.2.4</p>
<p>Zhang, Z.Y., Shao, Q.Y., Li, X., Chen, W.H., Liang, J.D., Han, Y.F., Huang, J.Z. & Liang, Z.Q. (2021) Culturable fungi from urban soils in China I: description of 10 new taxa. <em>Microbiology Spectrum</em> 9: e00867-21. https://doi.org/10.1128/Spectrum.00867-21</p>