Abstract
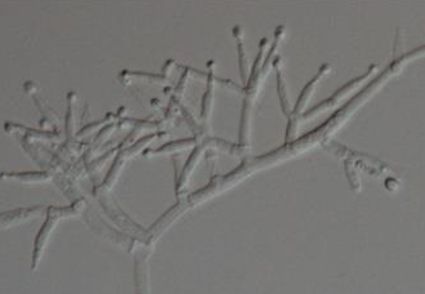
Trichoderma bombaxalis isolated from rhizosphere soils of five-year-old Lycium barbarum is described as a novel species. A combined approach is used to characterize T. bombaxalis including cultural and microscopic features, and phylogenetic analyses of the cascaded dataset of internal transcribed spacer (ITS) regions, RNA polymerase II subunit (RPB2), and the translation elongation factor 1-α gene (TEF1-α). The phylogeny reveals that T. bombaxalis belongs to section Hypocreanum, and is closely related to T. austriacum, T. eucorticoides, T. sulphureum, T. subsulphureum and T. victoriense, but represents a novel taxon. These fungi are characterized by often producing verticillium-like conidiophores and ellipsoidal conidia. Distinctions between the new species and its close relatives are compared and discussed. The ability of T. bombaxalis to degrade cellulose and hemicellulose is also estimated.
References
<p>Carbone, I & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. <em>Mycologia</em> 91: 553–556. https://doi.org/10.1080/00275514.1999.12061051</p>
<p>Castresana, J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. <em>Molecular Biology and Evolution</em> 17 (4): 540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334</p>
<p>Chaverri, P., Castlebury, L.A., Samuels, G.J. & David, M.G. (2003) Multilocus phylogenetic structure within the <em>Trichoderma harzianum</em>/<em>Hypocrea lixii</em> complex. <em>Molecular Phylogenetics and Evolution</em> 27 (2): 302–313. https://doi.org/10.1016/S1055-7903(02)00400-1</p>
<p>Chaverri, P., Branco-Rocha, F., Jaklitsch, W., Gazis, R., Degenkolb, T. & Samuels, G.J. (2015) Systematics of the <em>Trichoderma harzianum</em> species complex and the re-identification of commercial biocontrol strains. <em>Mycologia</em> 107 (3): 558–590. https://doi.org/10.3852/14-147</p>
<p>du Plessis, I.L., Druzhinina, I.S., Atanasova, L., Yarden, O. & Jacobs, K. (2018) The diversity of <em>Trichoderma</em> species from soil in South Africa, with five new additions. <em>Mycologia</em> 110: 559–583. https://doi.org/10.1080/00275514.2018.1463059</p>
<p>Edwards, J.C., Johnson, C., Santos-Medellin, E.L., Podishetty, N.K., Bhatnagar, S., Eisen, J.A. & Sundaresan, V. (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. <em>Proceedings of the National Academy of Sciences of the United States of America</em> 112: E911-20. https://doi.org/10.1073/pnas.1414592112</p>
<p>Ferreira, F.V., Herrmann-Andrade, A.M., Calabrese, C.D., Bello, F., Vazquez, D. & Musumeci, M.A. (2020) Effectiveness of <em>Trichoderma</em> strains isolated from the rhizosphere of citrus tree to control <em>Alternaria alternata</em>, <em>Colletotrichum gloeosporioides</em> and<em> Penicillium digitatum</em> A21 resistant to pyrimethanil in post-harvest oranges (<em>Citrus sinensis</em> L. (Osbeck)). <em>Journal of Applied Microbiology</em> 129: 712–727. https://doi.org/10.1111/jam.14657</p>
<p>Gu, X., Wang, R., Sun, Q., Wu, B. & Sun, J.Z. (2020) Four new species of <em>Trichoderma</em> in the <em>Harzianum </em>clade from northern China. <em>MycoKeys</em> 73: 109–132. https://doi.org/10.3897/mycokeys.73.51424</p>
<p>Guzman-Guzman, P., Porras-Troncoso, M.D., Olmedo-Monfil, V. & Herrera-Estrella, A. (2019) <em>Trichoderma </em>species: versatile plant symbionts. <em>Phytopathology</em> 109: 6–16. https://doi.org/10.1094/PHYTO-07-18-0218-RVW</p>
<p>Jaklitsch, W.M. (2009) European species of <em>Hypocrea</em> Part I. The green-spored species. <em>Studies in Mycology</em> 63: 1–91. https://doi.org/10.3114/sim.2009.63.01</p>
<p>Jaklitsch, W.M. (2011) European species of <em>Hypocrea</em> part II: species with hyaline ascospores. <em>Fungal Diversity</em> 48: 1–250. https://doi.org/10.1007/s13225-011-0088-y</p>
<p>Jaklitsch, W.M., Komon, M., Kubicek, C.P. & Druzhinina, I.S. (2017) <em>Hypocrea voglmayrii </em>sp. nov. from the Austrian Alps represents a new phylogenetic clade in<em> Hypocrea</em>/<em>Trichoderma</em>. <em>Mycologia</em> 97: 1365–1378. http://doi.org/10.1080/15572536.2006.11832743</p>
<p>Jang, S., Jang, Y., Kim, C.W., Lee, H., Hong, J.H., Heo, Y.M., Lee, Y.M., Lee, D.W., Lee, H.B. & Kim, J.J. (2017) Five new records of soil-derived <em>Trichoderma</em> in Korea: <em>T. albolutescens</em>, <em>T. asperelloides, T. orientale</em>, <em>T. spirale</em>, and <em>T. tomentosum</em>. <em>Mycobiology</em> 45: 1–8. https://doi.org/10.5941/MYCO.2017.45.1.1</p>
<p>Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. <em>Molecular Biology and Evolution </em>30: 772–780. https://doi.org/10.1093/molbev/mst010</p>
<p>Liu, B., Ji, S., Zhang, H., Wang, Y. & Liu, Z. (2020) Isolation of<em> Trichoderma</em> in the rhizosphere soil of <em>Syringa oblata</em> from Harbin and their biocontrol and growth promotion function. <em>Microbiological Research </em>235: 126445. https://doi.org/10.1016/j.micres.2020.126445</p>
<p>Liu, J.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. <em>Molecular Biology and Evolut</em>ion 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092</p>
<p>Na, X.F, Ma, S., Ma, C., Liu, Z., Xu, P., Zhu, H., Liang, W. & Kardol, P. (2021) <em>Lycium barbarum </em>L. (goji berry) monocropping causes microbial diversity loss and induces <em>Fusarium </em>spp<em>. </em>enrichment at distinct soil layers. <em>Applied Soil Ecology</em> 168: 104–107. https://doi.org/10.1016/j.apsoil.2021.104107</p>
<p>Nandini, B., Puttaswamy, H., Saini, R.K., Prakash, H.S. & Geetha, N. (2021) Trichovariability in rhizosphere soil samples and their biocontrol potential against downy mildew pathogen in pearl millet. <em>Scientifc Reports</em> 11: 9517. https://doi.org/10.1038/s41598-021-89061-2</p>
<p>Nylander, J.A.A. (2004) MrModeltest v2. Program distributed by the author. Available from: https://github.com/nylander/MrModeltest2</p>
<p>Overton, B.E., Stewart, E.L. & Geiser, D.M. (2006) Taxonomy and phylogenetic relationships of nine species of <em>Hypocrea</em> with an amorphs assignable to <em>Trichoderma</em> section <em>Hypocreanum</em>. <em>Studies in Mycology</em> 56: 39–65. https://doi.org/10.3114/sim.2006.56.02</p>
<p>Peciulyte, A., Anasontzis, G.E., Karlström, K., Larsson, P.T. & Olsson, L. (2014) Morphology and enzyme production of <em>Trichoderma reesei</em> Rut C-30 are affected by the physical and structural characteristics of cellulosic substrates. <em>Fungal Genetics and Biology</em> 72: 64–72. https://doi.org/10.1016/j.fgb.2014.07.011</p>
<p>Rambaut, A. (2012) FigTree version 1.4.0. Program distributed by the author. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 18 May 2022)</p>
<p>Riley, D. & Barber, S.A. (1970) Salt accumulation at the soybean (<em>Glycine max</em> ( L. ) Merr.) root-soil interface. <em>Soil Science Society of America Journal</em> 34 (1): 154–155. https://doi.org/10.2136/sssaj1970.03615995003400010042x</p>
<p>Ronquist, F., Teslenko, M., Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. <em>Systematic Biology</em> 61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Ryu, S.M., Lee, H.M., Song, E.G., Seo, Y.H., Lee, J., Guo, Y., Kim, B.S., Kim, J.J., Hong, J.S., Ryu, K.H. & Lee, D. (2017) Antiviral activities of trichothecenes isolated from <em>Trichoderma albolutescens</em> against pepper mottle virus. <em>Journal of Agricultural and Food Chemistry</em> 65: 4273–4279. https://doi.org/10.1021/acs.jafc.7b01028</p>
<p>Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. <em>Bioinformatics</em> 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446</p>
<p>Strakowska, J., Błaszczyk, L. & Chełkowski, J. (2014) The significance of cellulolytic enzymes produced by <em>Trichoderma</em> in opportunistic lifestyle of this fungus. <em>Journal of Basic Microbiology</em> 54: S2–S13. https://doi.org/10.1002/jobm.201300821</p>
<p>Swain, H., Adak, T., Mukherjee, A.K., Mukherjee, P.K., Bhattacharyya, P., Behera, S., Bagchi, T.B., Patro, R., Khandual, A., Bag, M.K., Dangar, T.K. & Jena, M. (2018) Novel <em>Trichoderma</em> strains isolated from tree barks as potential biocontrol agents and biofertilizers for direct seeded rice. <em>Microbiological Research</em> 214: 83–90. https://doi.org/10.1016/j.micres.2018.05.015</p>
<p>White, T.J., Bruns, T., Lee, S. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. <em>In</em>: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols: a Giude to Methods and Application. Academic Press, San Diego, USA, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</p>
<p>Zhang, J.H., Li, M., Jia, K.L., Zheng, G.Q. & Long, X.E. (2018) Seasonal variation rather than stand age determines bacterial diversity in the rhizosphere of wolfberry (<em>Lycium barbarum </em>L<em>.</em>) associated with soil degradation. <em>Journal of Soils and Sediments</em> 18: 1518–1529. https://doi.org/10.1007/s11368-017-1854-6</p>
<p>Zhang, R. & Wang, D. (2012) <em>Trichoderma </em>spp. from rhizosphere soil and their antagonism against <em>Fusarium sambucinum</em>. <em>African Journal of Biotechnology</em> 11 (18): 4180–4186. https://dx.doi.org/10.5897/ajb11.3426</p>
<p>Zhang, Y.J., Zhang, S., Liu, X.Z., Wen, H.A. & Wang, M. (2010) A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. <em>Letters in Applied Microbiology </em>51: 114–118. https://doi.org/10.1111/j.1472-765X.2010.02867.x</p>