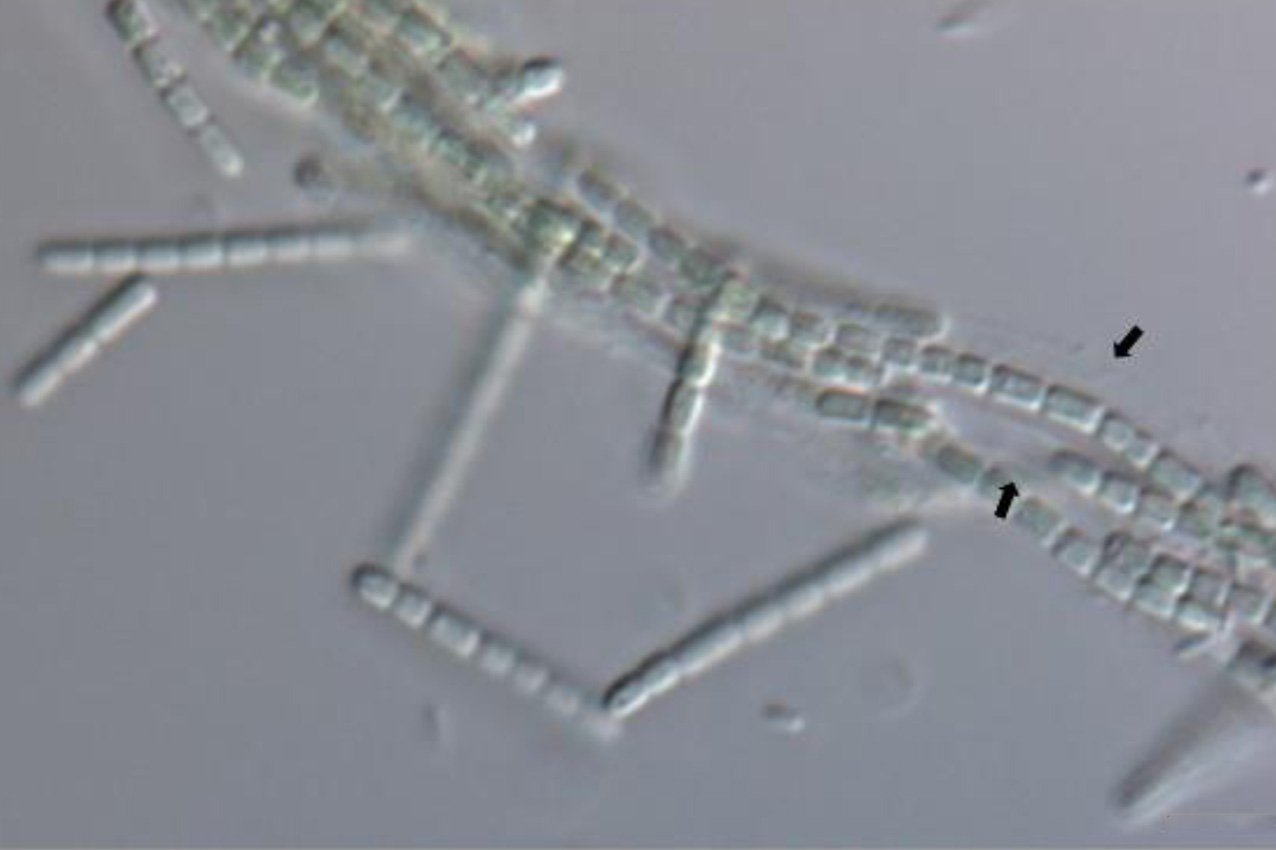
Abstract
The collection of Pinocchia daecheonga sp. nov. was performed at the Daecheong Lake in Korea, five strains of which with four clones were investigated through light microscopy, transmission electron microscopy and molecular data comprising the 16S rRNA to 23S rRNA gene. P. daecheonga differed from type species P. polymorpha by absence of the polar aerotopes. Investigated stains and four clones of P. daecheonga turned out to be a sister clade to the P. polymorpha according to the phylogenetic analysis of 16S rRNA gene. In addition, the Pinocchia was clustered with the family Leptolyngbyaceae members, genera Leptothoe and Leptoelongatus. 16S–23S rRNA intergenic transcribed spacer (ITS) region of P. daecheonga was found to be substantially distinct to P. polymorpha in terms of the secondary structure and nucleotide sequence composition, which concludes that the Pinocchia daecheonga isolated from the Daecheong Lake is a unique species due to differences in morphology and genetic traits compared to the relative P. polymorpha.
References
<p>Becerra-Absalón, I., Johansen, J.R., Muñoz-Martín, M.A. & Montejano, G. (2018) <em>Chroakolemma</em> gen. nov. (Leptolyngbyaceae, Cyanobacteria) from soil biocrusts in the semi-desert Central Region of Mexico. <em>Phytotaxa</em> 367: 201–218. https://doi.org/10.11646/phytotaxa.367.3.1</p>
<p>Casamatta, D.A., Johansen, J.R., Vis, M.L. & Broadwater, S.T. (2005) Molecular and morphological characterization of ten polar and near-polar strains within the Oscillatoriales (Cyanobacteria). <em>Journal of Phycology</em> 41: 421–438. https://doi.org/10.1111/j.1529-8817.2005.04062.x</p>
<p>Chakraborty, S., Maruthanayagam, V., Achari, A., Pramanik, A., Jaisankar, P. & Mukherjee, J. (2019) <em>Euryhalinema mangrovii</em> gen. nov., <em>sp. nov.</em> and <em>Leptoelongatus litoralis</em> gen. nov., <em>sp. nov.</em> (Leptolyngbyaceae) isolated from an Indian mangrove forest. <em>Phytotaxa</em> 422: 58–74. https://doi.org/10.11646/phytotaxa.422.1.4</p>
<p>Chun, S.-J., Cui, Y., Jin, C., Cho, A.-R., Wong, S.-K., Lee, H.-G., Oh, H.-M. & Ahn, C.-Y. (2020) <em>Paraconexibacter algicola</em> gen. nov., <em>sp. nov.</em>, a novel actinobacterium isolated from a eutrophic lake during the end of cyanobacterial harmful algal blooms, and proposal of Paraconexibacteraceae fam. nov. in the order Solirubrobacterales. <em>International Journal of Systematic and Evolutionary Microbiology</em> 70: 915–922. https://doi.org/10.1099/ijsem.0.003846</p>
<p>Dadheech, P.K., Mahmoud, H., Kotut, K. & Krienitz, L. (2012) <em>Haloleptolyngbya</em> <em>alcalis</em> <em>gen. et sp. nov.</em>, a new filamentous cyanobacterium from the soda lake Nakuru, Kenya. <em>Hydrobiologia</em> 691: 269–283. https://doi.org/10.1007/s10750-012-1080-6</p>
<p>Davydov, D., Shalygin, S. & Vilnet, A. (2020) New cyanobacterium <em>Nodosilinea svalbardensis</em> <em>sp. nov.</em> (Prochlorotrichaceae, Synechococcales) isolated from alluvium in Mimer river valley of the Svalbard archipelago. <em>Phytotaxa</em> 442: 61–79. https://doi.org/10.11646/phytotaxa.442.2.2</p>
<p>Dvořák, P., Jahodářová, E., Hašler, P., Gusev, E. & Poulíčková, A. (2015) A new tropical cyanobacterium <em>Pinocchia</em> <em>polymorpha</em> <em>gen. et sp. nov.</em> derived from the genus <em>Pseudanabaena</em>. <em>Fottea</em> 15: 113–120. https://doi.org/10.5507/fot.2015.010</p>
<p>Engene, N. & Tronholm, A. (2019) <em>Moorena</em> gen. nov., a valid name for “<em>Moorea</em> Engene & al.” nom. inval. Oscillatoriaceae, Cyanobacteria). <em>Notulae Algarum</em> 122: 1–2.</p>
<p>Gomont, M. (1892) Monographie des Oscillariées (Nostocacées Homocystées). Deuxième partie. -Lyngbyées. <em>Annales des Sciences Naturelles, Botanique, Série</em> 7 15: 263–368; 16: 91–264.</p>
<p>Guiry, M.D. & Guiry, G.M. (2021) AlgaeBase. World-wide electronic publication. National University of Ireland, Galway. Available from: http://www.algaebase.org/ (accessed 1 February 2021)</p>
<p>Hall, T.A. (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. <em>Nucleic Acids Symposia Series</em> 41: 95–98.</p>
<p>Hrouzek, P., Lukešová, A., Mareš, J. & Ventura, S. (2013) Description of the cyanobacterial genus <em>Desmonostoc</em> gen. nov. including <em>D</em>. <em>muscorum</em> <em>comb. nov.</em> as a distinct, phylogenetically coherent taxon related to the genus <em>Nostoc</em>. <em>Fottea</em> 13: 201–213. http://dx.doi.org/10.5507/fot.2013.016</p>
<p>Jahodářová, E., Dvořák, P., Hašler, P., Holušová, K. & Poulíčková, A. (2017) <em>Elainella</em> gen. nov.: a new tropical cyanobacterium characterized using a complex genomic approach. <em>European Journal of Phycology</em> 53: 39–51. https://doi.org/10.1080/09670262.2017.1362591</p>
<p>Johansen, J.R., Kovacik, L., Casamatta, D.A., Fučiková, K. & Kaštovský, J. (2011) Utility of 16S–23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: <em>Leptolyngbya</em> <em>corticola</em> <em>sp. nov.</em> (Pseudanabanaceae, Cyanobacteria). <em>Nova Hedwigia</em> 92: 283–302. https://doi.org/10.1127/0029-5035/2011/0092-0283</p>
<p>Kim, J.-H., Jeong, M.S., Kim, D.-Y., Her, S. & Wie, M.-B. (2015) Zinc oxide nanoparticles induce lipoxygenase-mediated apoptosis and necrosis in human neuroblastoma SH-SY5Y cells. <em>Neurochemistry International </em>90: 204–214. https://doi.org/10.1016/j.neuint.2015.09.002</p>
<p>Komárek, J. (2016) A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. <em>European Journal of Phycology</em> 51: 346–353. https://doi.org/10.1080/09670262.2016.1163738</p>
<p> </p>
<p>Komárek, J. (2018) Several problems of the polyphasic approach in the modern cyanobacterial system. <em>Hydrobiologia</em> 811: 7–17. https://doi.org/10.1007/s10750-017-3379-9</p>
<p>Komárek, J. & Anagnostidis, K. (2005) Cyanoprokaryota. 2. Teil: Oscillatoriales.<em> In: </em>Büdel, B., Krienitz, L., Gärtner, G. & Schagerl, M. (Eds.) <em>Süβwasserflora von Mitteleuropa, vol. </em>19/2. Elsevier, Spektrum, Heidelberg, pp. 1–759.</p>
<p>Komárek, J. & Lukavsky, J. (1988) <em>Arthronema</em>, a new cyanophyte genus from Afro-Asian deserts. <em>Archiv für Hydrobiologie, Supplement</em> 80: 249–267.</p>
<p>Komárek, J., Kaštovský, J., Mareš, J. & Johansen, J.R. (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. <em>Preslia</em> 86: 295–335.</p>
<p>Konstantinou, D., Voultsiadou, E., Panteris, E., Zervou, S.K., Hiskia, A. & Gkelis, S. (2019) <em>Leptothoe</em>, a new genus of marine cyanobacteria (Synechococcales) and three new species associated with sponges from the Aegean Sea. <em>Journal of Phycology</em> 55: 882–897. https://doi.org/10.1111/jpy.12866</p>
<p>Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. <em>Molecular Biology and Evolution</em> 33: 1870–1874. https://doi.org/10.1093/molbev/msw054</p>
<p>Lauterborn, H. (1915) Die sapropelische Lebewelt. Ein Beitrag zur biologie des Faulschlamms natürlicher Gewässer. <em>Verhandlungen des Naturhistorischen-Medizinischen Vereins zu Heidelberg</em>, 13: 395–481.</p>
<p>Lee, N.-J., Seo, Y.-S., Ki, J.-S. & Lee, O.-M. (2019) A study of newly recorded genus and species for aerial cyanobacteria <em>Wilmottia</em> <em>murrayi</em> (Oscillatoriales, Cyanobacteria) in Korea. <em>Korean Journal of Environmental Biology</em> 37: 260–267. https://doi.org/10.11626/KJEB.2019.37.3.260</p>
<p>Lee, N-J., Seo, Y., Ki, J.-S. & Lee, O.-M. (2020) Morphology and molecular description of <em>Wilmottia koreana</em> <em>sp. nov.</em> (Oscillatoriales, Cyanobacteria) isolated from the Republic of Korea. <em>Phytotaxa</em> 447: 237–251. https://doi.org/10.11646/phytotaxa.447.4.2</p>
<p>Li, X. & Li, R. (2016) <em>Limnolyngbya circumcreta</em> gen. & <em>comb. nov.</em> (Synechococcales, Cyanobacteria) with three geographical (provincial) genotypes in China. <em>Phycologia</em> 55: 478–491. https://doi.org/10.2216/15-149.1</p>
<p>Mai, T., Johansen, J.R., Pietrasiak, N., Bohunická, M. & Martin, M.P. (2018) Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. <em>Phytotaxa</em> 365: 1–59. https://doi.org/10.11646/phytotaxa.365.1.1</p>
<p>Miscoe, L.H., Johansen, J.R., Vaccarino, M.A., Pietrasiak, N. & Sherwood, A.R. (2016) Novel cyanobacteria from caves on Kauai, Hawaii. <em>Bibliotheca Phycologica</em> 120: 75–152.</p>
<p>Osario-Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Kováčik, L., Martin, M.P & Johansen, J.R. (2014) Seven new species of <em>Oculatella</em> (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. <em>European Journal of Phycology</em> 49: 450–470. https://doi.org/10.1080/09670262.2014.976843</p>
<p>Page, R.D.M. (1996) TreeView: An application to display phylogenetic trees on personal computers. <em>Bioinformatics</em> 12: 357–358.</p>
<p>Perkerson III, R.B., Johansen, J.R., Kováčik, L., Brand, J., Kaštovský, J. & Casamatta, D.A. (2011) A unique pseudanabaenalean (Cyanobacteria) genus <em>Nodosilinea</em> gen. nov. based on morphological and molecular data. <em>Journal of Phycology</em> 47: 1397–1412. https://doi.org/10.1111/j.1529-8817.2011.01077.x</p>
<p>Pietrasiak, N., Osorio-Santos, K., Shalygin, S., Martin, M.P. & Johansen, J.R. (2019) When is a lineage a species? a case study in <em>Myxacorys</em> gen. nov. (Synechococcales: Cyanobacteria) with the description of two new species from the Americas. <em>Journal of Phycology</em> 55: 976–996. https://doi.org/10.1111/jpy.12897</p>
<p>Raabová, L., Kovacik, L., Elster, J. & Strunecký, O. (2019) Review of the genus <em>Phormidesmis</em> (Cyanobacteria) based on environmental, morphological, and molecular data with description of a new genus <em>Leptodesmis</em>. <em>Phytotaxa</em> 395: 1–16. https://doi.org/10.11646/phytotaxa.395.1.1</p>
<p>Richards, E., Reichardt, M. & Rogers, S. (2003) Preparation of genomic DNA from plant tissue.<em> In: </em>Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. (Eds.) <em>Current protocols in molecular biology</em>. John Wiley and Sons, Inc., New York, pp. 2.3.1–2.3.7.</p>
<p>Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inferenceunder mixed models. <em>Bioinformatics</em> 19: 1572–1574. http://dx.doi.org/10.1093/bioinformatics/btg180</p>
<p>Siegesmund, M.A., Johansen, J.R., Karsten, U. & Friedl, T. (2008) <em>Coleofasciculus</em> gen. nov. (Cyanobacteria): morphological and molecular criteria for revision of the genus <em>Microcoleus</em> Gomont. <em>Journal of Phycology</em> 44: 1572–1585. https://doi.org/10.1111/j.1529-8817.2008.00604.x</p>
<p>Song, G., Jiang, Y. & Li, R. (2015) <em>Scytolyngbya</em> <em>timoleontis</em>, <em>gen. et sp. nov.</em> (Leptolyngbyaceae, Cyanobacteria): a novel false branching cyanobacteria from China. <em>Phytotaxa</em> 224: 72–84. https://doi.org/10.11646/phytotaxa.224.1.5</p>
<p>Stackebrandt, E. & Goebel, B.M. (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. <em>International Journal of Systematic Bacteriology</em> 44: 846–849.</p>
<p>Stanier, R.Y., Kunisawa, R., Mandel, M. & Cohenbaz, G. (1971) Purification and properties of unicellular blue-green algae (Order Chroococcales). <em>Bacteriological Reviews</em> 35: 171–205.</p>
<p>Taton, A., Wilmotte, A., Šmarda, J., Elster, J. & Komárek, J. (2011) <em>Plectolyngbya</em> <em>hodgsonii</em>: a novel filamentous cyanobacterium from Antarctic lakes. <em>Polar Biology</em> 34: 181–191. http://dx.doi.org/10.1007/s00300-010-0868-y</p>
<p>Vaz, M.G.M.V., Bonaldo-Genuário, D., Dini-Andreote, A.P., Silva-Malone, C.F., Sant’-Anna, C.L., Barbiero, L. & Fátima-Fiore, M. (2015) <em>Pantanalinema</em> gen. nov. and <em>Alkalinema</em> gen. nov.: novel pseudanabaenacean genera (Cyanobacteria) isolated from saline–alkaline lakes. <em>International Journal of Systematic and Evolutionary Microbiology</em> 65: 298–308. https://doi.org/10.1099/ijs.0.070110-0</p>
<p>Zammit, G., Billi, D. & Albertano, P. (2012) The subaerophytic cyanobacterium <em>Oculatella</em> <em>subterranea</em> (Oscillatoriales, Cyanophyceae) <em>gen. et sp. nov.</em>: a cytomorphological and molecular description. <em>European Journal of Phycology</em> 47: 341–354. http://dx.doi.org/ 10.1080/09670262.2012.717106</p>
<p>Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. <em>Nucleic Acids Research</em> 31: 3406–3415. http://dx.doi.org/10.1093/nar/gkg595</p>