Abstract
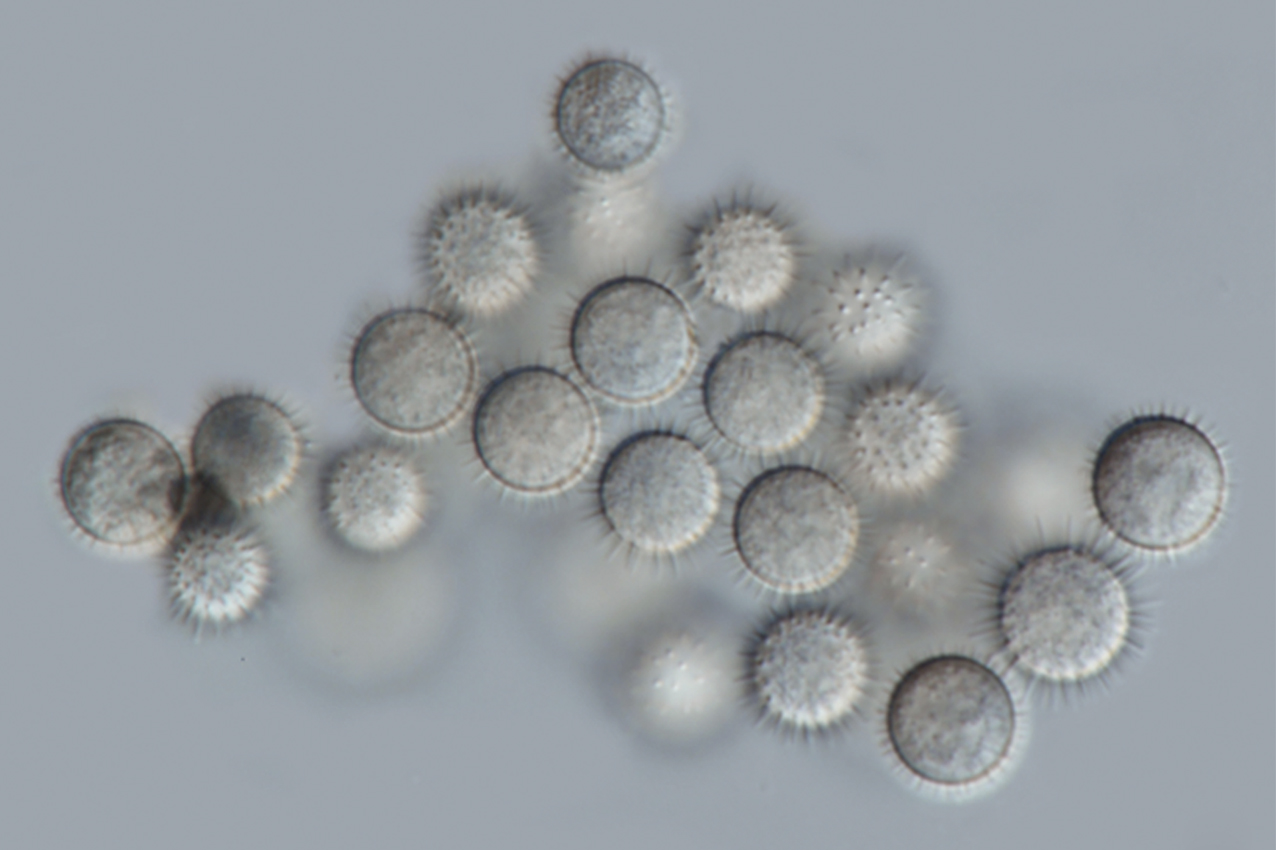
A new endophytic fungus, described herein as Cunninghamella saisamornae, was isolated from soil collected from Chiang Mai Province, Thailand. This species was distinguished from the previously described Cunninghamella species by the presence of a chlamydospore formation, the lack of a zygospore stage and a maximum growth temperature of 45°C. Multi-gene phylogenetic analysis of internal transcribed spacers (ITS) and large subunit (LSU) of the nuclear ribosomal DNA (rDNA), along with the translation elongation factor 1-alpha (tef-1) genes, support the finding that C. saisamornae is distinct from other species within the genus Cunninghamella. A full description, color photographs, illustrations and a phylogenetic tree showing the position of C. saisamornae are provided.
References
<p>Alfaro, M.E., Zoller, S. & Lutzoni, F. (2003) Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. <em>Molecular Biology and Evolution </em>20: 255–266. https://doi.org/10.1093/molbev/msg028</p>
<p>Alves, A.L., de Souza, C.A., de Oliveira, R.J., Cordeiro, T.R. & de Santiago, A.L. (2017) <em>Cunninghamella clavata </em>from Brazil: a new record for the western hemisphere. <em>Mycotaxon</em> 132: 381–389.</p>
<p>Baijai, U. & Mehrotra, B.S. (1980) The genus <em>Cunninghamella</em> – a reassessment. <em>Sydowia</em> 33: 1–13.</p>
<p>Bragulat, M.R., Castella, G., Isidoro-Ayza, M., Domingo, M. & Cabanes, F.J. (2017) Characterization and phylogenetic analysis of a <em>Cunninghamella bertholletiae</em> isolate from a bottlenose dolphin (<em>Tursiops truncatus</em>). <em>Revista Iberoamericana de Micología</em> 34: 215–219. http://dx.doi.org/10.1016/j.riam.2017.03.002</p>
<p>Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. <em>Nature Methods </em>9: 772. https://doi.org/10.1038/nmeth.2109</p>
<p>Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. <em>Nucleic Acids Research </em>32: 1792–1797. https://doi.org/10.1093/nar/gkh340</p>
<p>Felsenstein, J. (1985) Confidence intervals on phylogenetics: an approach using bootstrap. <em>Evolution </em>39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x</p>
<p>Guo, J., Wang, H., Liu, D., Zhang, J.N., Zhao, Y.H., Liu, T.X. & Xin, Z.H. (2015) Isolation of <em>Cunninghamella bigelovii</em> <em>sp. nov.</em> CGMCC 8094 as a new endophytic oleaginous fungus from <em>Salicornia bigelovii</em>. <em>Mycological Progress</em> 14: 11 https://doi.org/10.1007/s11557-015-1029-z</p>
<p>Hillis, D.M. & Bull, J.J. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. <em>Systematic Biology </em>42: 182–192.</p>
<p>Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., McKenzie, E.H.C., Jones, E.B.G., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q., Abdel-Aziz, F.A., Abdel-Wahab, M., Banmai, S., Chomnunti, P., Cui, B.K., Daranagama, D.A., Das, K., Dayarathne, M.C., De Silva, N.I., Dissanayake, A.J., Doilom, M., Ekanayaka, A.H., Gibertoni, T.B., Góes-Neto, A., Huang, S.K., Jayasiri, S.C., Jayawardena, R.S., Konta, S., Lee, H.B., Li, W.J., Lin, C.G., Liu, J.K., Lu, Y.Z., Luo, Z.L., Manawasinghe, I.S., Manimohan, P., Mapook, A., Niskanen, T., Norphanphoun, C., Papizadeh, M., Perera, R.H., Phukhamsakda, C., Richter, C., De Santiago, A.L.C.M., Drechsler-Santos, E.R., Senanayake, I.C. & Tanaka, K. (2016) Fungal diversity notes 367-490: taxonomic and phylogenetic contributions to fungal taxa. <em>Fungal Diversity </em>80: 1–270. https://doi.org/10.1007/s13225-016-0373-x</p>
<p>Liu, X.Y., Huang, H. & Zheng, R.Y. (2001) Relationships within <em>Cunninghamella </em>based on sequence analysis of its rDNA. <em>Mycotaxon</em> 80: 77–95.</p>
<p>Liu, C.W., Liou, G.Y. & Chien, C.Y. (2005) New records of the genus <em>Cunninghamella </em>(Mucorales) in Taiwan. <em>Fungal Science</em> 20: 1–9.</p>
<p>Matruchot, L. (1903) Une Mucorinéepurementconidienne, <em>Cunninghamella Africana</em>. <em>Annales Mycologici </em>1: 45–60.</p>
<p>Nilsson, R.H., Kristiansson, E., Ryberg, M., Hallenberg, N. & Larsson, K. (2008) Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. <em>Evolutionary Bioinformatics</em> 4: 193–201. https://doi.org/10.4137/ebo.s653</p>
<p>Nguyen, T.T.T., Choi, Y.J. & Lee, H.B. (2017) Zygomycete fungi in Korea: <em>Cunninghamella bertholletiae</em>, <em>Cunninghamella echinulata</em>, and <em>Cunninghamella elegans</em>. <em>Mycobiology</em> 45: 318–326. https://doi.org/10.5941/MYCO.2017.45.4.318</p>
<p>O’Donnell, K., Lutzoni, F.M., Ward, T.J. & Benny, G.L. (2001) Evolutionary relationships among mucoralean fungi Zygomycota: Evidence for family polyphyly on a large scale. <em>Mycologia </em>93: 286–297. https://doi.org/10.1080/00275514.2001.12063160</p>
<p>Reed, M.D.A., Body, P.B., Austin, M.D.M.B. & Jr Frierson, M.D.H. (1988) <em>Cunninghamella bertholletiae</em> and <em>Pneumocystis carinii</em> pneumonia as a fatal complication of chronic lymphocytic leukemia. <em>Human Pathology</em> 19: 1470–1472.</p>
<p>Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. <em>Systematic Biology</em> 61: 539–542. https://doi.org/10.1093/sysbio/sys029</p>
<p>Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. <em>Bioinformatics</em> 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446</p>
<p>Tamura, K., Stecher, G., Petersen, D., Filipsky, A. & Kumar, S. (2013) MEGE 6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolutionary 30: 2725–2729. https://doi.org/10.1093/molbev/mst197</p>
<p>Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several <em>Cryptococcus </em>species. <em>Journal of Bacteriology </em>172: 4238–4246. https://doi.org/10.1128/JB.172.8.4238-4246.1990</p>
<p>Walther, G., Pawłowska, J., Alastruey-Izquierdo, A., Wrzosek, M., Rodriguez-Tudela, J.L., Dolatabadi, S., Chakrabarti, A. & de Hoog, GS. (2013) DNA barcoding in Mucorales: an inventory of biodiversity. <em>Persoonia</em> 30: 11–47. https://doi.org/10.3767/003158513X665070. </p>
<p>White, T.J., Burns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics.<em> In: </em>Innis, M.A., Gelfand, D.H., Sninsky, J.J. & Whitish, T.J. (Eds.) <em>PCR Protocols, a Guide to Methods and Applications.</em> Academic Press, San Diego. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</p>
<p>Yu, J., Walther, G., Van Diepeningen, A.D., Gerrits Van Den Ende, A.H.G., Li, R.Y., Moussa, T.A.A., Almaghrabi, O.A. & De Hoog, G.S. (2015) DNA barcoding of clinically relevant <em>Cunninghamella</em> species. <em>Medical mycology</em> 53: 99–106. https://doi.org/10.1093/mmy/myu079</p>
<p>Zhang, Z.Y., Zhao, Y.X., Shen, X., Chen, W.H., Han, Y.F., Huang, J.Z. & Liang, Z.Q. (2020) Molecular phylogeny and morphology of <em>Cunninghamella guizhouensis sp. nov.</em> (Cunninghamellaceae, Mucorales), from soil in Guizhou, China. <em>Phytotaxa</em> 455: 31–39. https://doi.org/10.11646/phytotaxa.455.1.4</p>
<p>Zheng, R.Y. & Chen, G.Q. (2001) A monograph of <em>Cunninghamella</em>. <em>Mycotaxon</em> 80: 1–75.</p>