Abstract
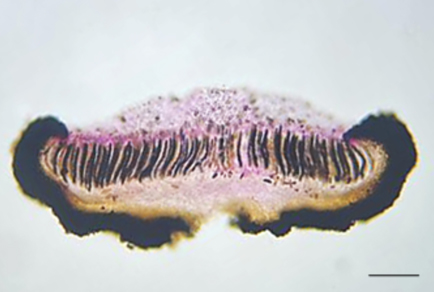
The genus Rhytidhysteron is ecologically diverse, cosmopolitan group of Dothideomycetes characterized by apothecioid ascomata, in which the margin folds inwards, covers the disc incompletely in dry conditions, unfolds outwards exposing the disc by a lenticular or irregular opening when hydrated, bitunicate, cylindrical asci, and hyaline to reddish brown, ellipsoidal to fusiform, 1–3 septate ascospores. This study was conducted on five Rhytidhysteron specimens isolated from dead wood pieces in urban forest habitats (Colombo District, Sri Lanka). Detailed micro and macro morphological examinations and phylogenetic analyses inferred from ITS, LSU, and TEF1 were used to position the five specimens compared with the extant Rhytidhysteron taxa. Based on morphological and phylogenetic evidence, they were identified as R. rufulum, R. neorufulum, R. cf. bruguiereae and a novel species, R. constrictiseptum; altogether providing first records for three Rhytidhysteron species in Sri Lanka, except R. neorufulum.
References
- Alster, C.J., Allison, S.D., Johnson, N.G., Glassman, S.I. & Treseder, K.K. (2021) Phenotypic plasticity of fungal traits in response to moisture and temperature. ISME Communications 1 (1): 43. https://doi.org/10.1038/s43705-021-00045-9
- Anahosur, K.H. (1971) Ascomycetes of Coorg (India) II. Sydowia 24: 177–182.
- Atti della Reale accademia delle scienze fisiche e matematiche. (1879) Reale Accademia delle scienze fisiche e matematiche di NapoliSocietà reale di Napoli. Vol. 8. Tip. della R. Accademia delle scienze fisiche et matematiche Napoli, pp. 1863–1935.
- Boehm, E.W.A., Mugambi, G.K., Miller, A.N., Huhndorf, S.M., Marincowitz, S., Spatafora, J.W. & Schoch, C.L. (2009a) A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Studies in Mycology 64: 49–83. https://doi.org/10.3114/sim.2009.64.03
- Boehm, E.W.A., Schoch, C.L. & Spatafora, J.W. (2009b) On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycological Research 113: 461–479. https://doi.org/10.1016/j.mycres.2008.12.001
- Camino-Vilaró, M., Castro-Hernández, L., Abreu-Herrera, Y., Mena-Portales, J. & Cantillo-Pérez, T. (2019) Fungi associated with invasive plant species in Cuba. Phytotaxa 419 (3): 239–267. https://doi.org/10.11646/phytotaxa.419.3.1
- Chevallier, F.F. (1826) Flore générale des environs de Paris, Vol. I. Ferra LibrairieEditeur, Paris, France.
- Cobos Villagrán, A., Hernández Rodríguez, C.H., Valenzuela, R., Villa-Tanaca, L., Calvillo-Medina, R.P., Mateo-Cid, L.E., Martínez-Pineda, M. & Raymundo, T. (2020) El género Rhytidhysteron (Dothideomycetes, Ascomycota) en México. Acta Botanica Mexicana 127. https://doi.org/10.21829/abm127.2020.1675
- Cobos-Villagrán, A., Raymundo, T., Calvillo-Medina, R.P. & Valenzuela, R. (2021a) Rhytidhysteron mexicanum sp. Nov. (Dothideomycetes, Ascomycota) from the Sierra of Guadalupe, Trans Mexican Volcanic Belt. Phytotaxa 479 (3): 275–286. https://doi.org/10.11646/phytotaxa.479.3.4
- Cobos-Villagrán, A., Valenzuela, R., Hernández-Rodríguez, C., Calvillo-Medina, R.P., Villa-Tanaca, L., Mateo-Cid, L.E., Pérez-Valdespino, A. & Martínez-González, C.R. & Raymundo, T. (2021) Three new species of Rhytidhysteron (Dothideomycetes, Ascomycota) from Mexico. MycoKeys 83: 123–144. https://doi.org/10.3897/mycokeys.83.68582
- Dayarathne, M.C., Jones, E.B.G., Maharachchikumbura, S.S.N., Devadatha, B., Sarma, V.V., Khongphinitbunjong, K., Chomnunti, P. & Hyde, K.D. (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11 (1): 1–188. https://doi.org/10.5943/mycosphere/11/1/1
- De Almeida, D.A.C., Gusmão, L.F.P. & Miller, A.N. (2014) A new genus and three new species of hysteriaceous ascomycetes from the semiarid region of Brazil. Phytotaxa 176 (1): 298–308. https://doi.org/10.11646/phytotaxa.176.1.28
- De Silva, N.I., Tennakoon, D.S., Thambugala, K.M., Karunarathna, S.C., Lumyong, S. & Hyde, K.D. (2020) Morphology and multigene phylogeny reveal a new species and a new record of Rhytidhysteron (Dothideomycetes, Ascomycota) from China. Asian Journal of Mycology 3 (1): 295–306. https://doi.org/10.5943/ajom/3/1/4
- De Silva, N.T., Thambugala, K.M. & Ekanayaka, A.H. (2025) Morpho-Molecular Diversity of Rhytidhysteron in Sri Lanka with Insights into Intraspecific Variation of R. rufulum. In: Proceedings of International Conference on Microbes and Microbial Technologies-2025. The Faculty of Technology, Rajarata University of Sri Lanka, pp. 86.
- Desai, B.G. & Pathak, V.N. (1970) A new Tryblidiella from India. Sydowia 24: 198–200.
- Doilom, M., Dissanayake, A.J., Wanasinghe, D.N., Boonmee, S., Liu, J.-K., Bhat, D.J., Taylor, J.E., Bahkali, A.H., McKenzie, E.H.C. & Hyde, K.D. (2017) Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Diversity 82 (1): 107–182. https://doi.org/10.1007/s13225-016-0368-7
- Du, T.Y., Dai, D.Q., Mapook, A., Lu, L., Stephenson, S.L., Suwannarach, N., Elgorban, A.M., Al-Rejaie, S., Karunarathna, S.C. & Tibpromma, S. (2023) Additions to Rhytidhysteron (Hysteriales, Dothideomycetes) in China. Journal of Fungi 9 (2): 148. https://doi.org/10.3390/jof9020148
- Ekanayaka, A.H., Ariyawansa, H.A., Hyde, K.D., Jones, E.B.G., Daranagama, D.A., Phillips, A.J.L., Hongsanan, S., Jayasiri, S.C. & Zhao, Q. (2017) DISCOMYCETES: The apothecial representatives of the phylum Ascomycota. Fungal Diversity 87 (1): 237–298. https://doi.org/10.1007/s13225-017-0389-x
- Francisco, C.S., Ma, X., Zwyssig, M.M., McDonald, B.A. & Palma-Guerrero, J. (2019) Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici. Scientific Reports 9 (1): 9642. https://doi.org/10.1038/s41598-019-45994-3
- Hall, T.A. (1998) BioEdit, a user-friendly biological sequence alignment editor and analysisprogrm for Windows95/98/NT. Nucleic Acids Symposium 41: 95–98.
- Hoang, D.T., Chernomor, O., von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 (8): 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Hyde, K.D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D.J., Jones, E.B.G., Liu, N.G., Abeywickrama, P.D., Mapook, A., Wei, D., Perera, R.H., Manawasinghe, I.S., Pem, D., Bundhun, D., Karunarathna, A., Ekanayaka, A.H., Bao, D.F., Li, J., Samarakoon, M.C., Chaiwan, N., Lin, C.G., Phutthacharoen, K., Zhang, S.N., Senanayake, I.C., Goonasekara, I.D., Tambugala, K.M., Phukhamsakda, C. & Tennakoon, D.S., Jiang, H.B., Yang, J., Zeng, M., Huanraluek, N., Liu, J.K., Wijesinghe, S.N., Tian, Q., Tibpromma, S., Brahmanage, R.S., Boonmee, S., Huang, S.K., Tiyagaraja, V., Lu, Y.Z., Jayawardena, R.S., Dong, W., Yang, E.F., Singh, S.K., Singh, S.M., Rana, S., Lad, S.S., Anand, G., Devadatha, B., Niranjan, M., Sarma, V.V., Liimatainen, K., Aguirre-Hudson, B., Niskanen, T., Overall, A., Alvarenga, R.L.M., Gibertoni, T.B., Pfliegler, W.P., Horváth, E., Imre, A., Alves, A.L., da Silva Santos, A.C., Tiago, P.V., Bulgakov, T.S., Wanasinghe, D.N., Bahkali, A.H., Doilom, M., Elgorban, A.M., Maharachchikumbura, S.S.N., Rajeshkumar, K.C., Haelewaters, D., Mortimer, P.E., Zhao, Q., Lumyong, S., Xu, J. & Sheng, J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. https://doi.org/10.1007/s13225-020-00439-5
- Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., McKenzie, E.H.C., Jones, E.B.G., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Banmai, S., Chomnunti, P., Cui, B.-K., Daranagama, D.A., Das, K., Dayarathne, M.C., De Silva, N.I. & Zhu, L. (2016) Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80 (1): 1–270. https://doi.org/10.1007/s13225-016-0373-x
- Hyde, K.D., Gareth Jones, E.B., Liu, J.K., Ariyawansa, H., Boehm, E., Boonmee, S., Braun, U., Chomnunti, P., Crous, P.W., Dai, D.Q., Diederich, P., Dissanayake, A., Doilom, M., Doveri, F., Hongsanan, S., Jayawardena, R., Lawrey, J.D., Li, Y.M., Liu, Y.Z., Lücking, R., Monkai, J., Muggia, L., Nelsen, M.P., Pang, K.L., Phookamsak, R., Senanayake, I.C., Shearer, C.A., Suetrong, S., Tanaka, K., Thambugala, K.M., Wijayawardene, N.N., Wikee, S., Wu, H.X., Zhang, Y., Aguirre-Hudson, B., Aisyah Alias, S., Aptroot, A., Bahkali, AH., Bezerra, J.L., Jayarama Bhat, D., Camporesi, E., Chukeatirote, E., Gueidan, C., Hawksworth, D.L., Hirayama, K., De Hoog, S., Kang, J.C., Knudsen, K., Li, W.J., Li, X.H., Liu, Z.Y., Mapook, A., McKenzie, E.H.C., Miller, A.N., Mortimer, P.E., Phillips, A.J.L., Raja, H.A., Scheuer, C., Schumm, F., Taylor, J.E., Tian, Q., Tibpromma, S., Wanasinghe, D.N., Wang, Y. & Xu, J.C. (2013) Families of Dothideomycetes. Fungal Diversity 63: 1–313. https://doi.org/10.1007/s13225-013-0263-4
- Hyde, K.D., Noorabadi, M.T., Thiyagaraja, V., He, M.Q., Johnston, P.R., Wijesinghe, S.N., Armand, A., Biketova, A.Y., Chethana, K.W.T., Erdogdu, M., Ge, Z.W., Groenewald, J.Z., Hongsanan, S., Kusan, I., Leontyev, D.V., Li, D.W., Lin, C.G., Liu, N.G., Maharachchikumburua, S.S.N., Matocec, N., May, T.W., McKenzie, E.H.C., Mesic, A., Perera, R.H., Phukhamsakda, C., Piqtek, M., Samarakoon, M.C., Selcuk, F., Senanayake, I.C., Tanney, J.B., Tian, Q., Vizzini, A., Wanasinghe, D.N., Wannasawang, N., Wijayawardene, N.N. & Zhao, R.L. (2024) The 20204 Outline of Fungi and Fungus-like taxa. Mycosphere 15 (1): 5146–6239. https://doi.org/10.5943/mycosphere/15/1/25
- Index Fungorum (2025) Index Fungorum. MycoBank and Index Fungorum Partnership. Available from: https://www.indexfungorum.org/names/names.asp (accessed 2 August 2025)
- Jayasiri, S., Hyde, K.D., Jones, E.B.G., Persoh, D., Camporesi, E. & Kang, J.C. (2018) Taxonomic novelties of hysteriform Dothideomycetes. Mycosphere 9 (4): 803–837. https://doi.org/10.5943/mycosphere/9/4/8
- Jayasiri, S.C., Hyde, K.D., Jones, E.B.G., McKenzie, E.H.C., Jeewon, R., Phillips, A.J.L., Bhat, D.J., Wanasinghe, D.N., Liu, J.K., Lu, Y.Z. & Kang, J.C. (2019) Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10 (1): 1–186. https://doi.org/10.5943/mycosphere/10/1/1
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7 (11): 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285
- Karsten, P. (1866) Enumeratio Fungorum et Myxomycetum in Lapponia orientale aestate 1861 lectorum. Notiser ur Sällskapets pro Fauna et Flora Fennica Förhandlingar 8: 193–224.
- Kumar, V., Cheewangkoon, R., Thambugala, K.M., Jones, G.E.B., Brahmanage, R.S., Doilom, M., Jeewon, R. & Hyde, K.D. (2019) Rhytidhysteron mangrovei (Hysteriaceae), a new species from mangroves in Phetchaburi Province, Thailand. Phytotaxa 401 (3): 166. https://doi.org/10.11646/phytotaxa.401.3.2
- Luttrell, E.S. (1965) Paraphysoids, pseudoparaphyses, and apical paraphyses. Transactions of the British Mycological Society 48 (1): 135–144. https://doi.org/10.1016/S0007-1536(65)80018-3
- Magnes, K.H. (1997) Weltmonographie der Triblidiaceae. Bibliotheca Mycologica 165: 1–177.
- Mapook, A., Hyde, K.D., McKenzie, E.H.C., Jones, E.B.G., Bhat, D.J., Jeewon, R., Stadler, M., Samarakoon, M.C., Malaithong, M., Tanunchai, B., Buscot, F., Wubet, T. & Purahong, W. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Diversity 101 (1): 1–175. https://doi.org/10.1007/s13225-020-00444-8
- Marian, I.M., Valdes, I.D., Hayes, R.D., LaButti, K., Duffy, K., Chovatia, M., Johnson, J., Ng, V., Lugones, L.G., Wosten, H.A.B., Grigoriev, I.V. & Ohm, R.A. (2024) High phenotypic and genotypic plasticity among strains of the mushroom-forming fungus Schizophyllum commune, Fungal Genetics and Biology 173: 103913. https://doi.org/10.1016/j.fgb.2024.103913
- Mudhigeti, N., Patnayak, R., Kalawat, U. & Yeddula, S.R.C. (2018) Subcutaneous Rhytidhysteron Infection: A Case Report from South India with Literature Review. Cureus 10 (4): e2406. https://doi.org/10.7759/cureus.2406
- Mugambi, G.K. & Huhndorf, S.M. (2009) Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi). Systematics and Biodiversity 7 (4): 453–464. https://doi.org/10.1017/S147720000999020X
- Murillo, C., Albertazzi, F.J., Carranza, J., Lumbsch, H.T. & Tamayo, G. (2009) Molecular data indicate that Rhytidhysteron rufulum (Ascomycetes, Patellariales) in Costa Rica consists of four distinct lineages corroborated by morphological and chemical characters. Mycological Research 113: 405–416. https://doi.org/10.1016/j.mycres.2008.09.003
- Niranjan, M., Tiwari, S., Baghela, A. & Sarva, V.V. (2018) New records of Ascomycetous fungi from Andaman Islands, India and their molecular sequence data. Current Research in Environmental & Applied Mycology 8 (3): 331–350. https://doi.org/10.5943/cream/8/3/5
- Peck, C.H. (1894) Report of the Botanist (1892). Annual Report on the New York State Museum of Natural History 46: 85–149.
- Pem, D., Jeewon, R., Chethana, K.W.T., Hongsanan, S., Doilom, M., Suwannarach, N. & Hyde, K.D. (2021) Species concepts of Dothideomycetes: Classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Diversity 109 (1): 283–319. https://doi.org/10.1007/s13225-021-00485-7
- Penzig, A.J.O. & Saccardo, P.A. (1897) Diagnoses fungorum novorum in insula Java collectorum. Ser. II. Malpighia 11: 491–530.
- Pudhom, K. & Teerawatananond, T. (2014) Rhytidenones A–F, Spirobisnaphthalenes from Rhytidhysteron sp. AS21B, an Endophytic Fungus. Journal of Natural Products 77 (8): 1962–1966. https://doi.org/10.1021/np500068y
- Ramírez-López, I., Rios, M.V. & Cano-Santana, Z. (2013) Phenotypic plasticity of the basidiomata of Thelephora sp. (Thelephoraceae) in tropical forest habitats, Revista de Biologia. Tropical 61 (1): 343–350. https://doi.org/10.15517/rbt.v61i1.11133
- Rambaut, A. & Drummond, A. (2009) Tracer v1.5. Available from: http://tree.bio.ed.ac.uk/software/tracer/ (accessed 10 January 2026)
- Rambaut, A. (2012) FigTree v1.4.0. Available from: http://tree.bio.ed.ac.uk/software/figtree (accessed 11 January 2026)
- Rannala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43 (3): 304–315. https://doi.org/10.1007/BF02338839
- Rashmi, M., Kushveer, J.S. & Sarma, V.V. (2019) A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10: 798–1079. https://doi.org/10.5943/mycosphere/10/1/19
- Ren, G.C., Wanasinghe, D.N., Jeewon, R., Monkai, J., Mortimer, P.E., Hyde, K.D., Xu, J.C. & Gui, H. (2022) Taxonomy and phylogeny of the novel Rhytidhysteron-like collections in the Greater Mekong Subregion. MycoKeys 86: 65–85. https://doi.org/10.3897/mycokeys.86.70668
- Samuels, F.E. & Muller, E. (1979) Rhytidhysteron rufulum and the genus Eutryblidiella. Life-history studies of Brazilian Ascomycetes. Sydowia 32: 277–292.
- Senanayake, I., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhum, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 11 (1): 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
- Senwanna, C., Suwannarach, N., Phookamsak, R. & Kumla, J., Hongsanan, S. (2023) New host and geographical records of Rhytidhysteron in nothern Thailand, and species synonymization. Phytotaxa 601 (2): 157–173. https://doi.org/10.11646/phytotaxa.601.2.3
- Sharma, M.P. & Rawla, G.S. (1986) Ascomycetes new to India – III. Nova Hedwigia 42: 81–90.
- Slepecky, R.A. & Starmer, W.T. (2009) Phenotypic plasticity in fungi: a review with observations on Aureobasidium pullulans. Mycologia 101 (6): 823–832. https://doi.org/10.3852/08-197
- Soto-Medina, E.A. & Lücking, R. (2017) A new species of Rhytidhysteron (Ascomycota: Patellariaceae) from Colombia, with a provisional working key to known species in the world. Revista de La Academia Colombiana de Ciencias Exactas, Físicas y Naturales 41 (158): 59–63. https://doi.org/10.18257/raccefyn.423
- Spegazzini, C. (1881) Fungi argentini additis nonnullis brasiliensibus montevideensibuaque. Pugillus Quartus (Continuacion). Anales de la Sociedad Cientifica Argentina 12: 174–189.
- Thambugala, K.M., Hyde, K.D., Eungwanichayapant, P.D. & Liu, Z.-Y. (2016) Additions to the Genus Rhytidhysteron in Hysteriaceae. Cryptogamie, Mycologie 37 (1): 99–116. https://doi.org/10.7872/crym/v37.iss1.2016.99
- Thambugala, K.M., Hyde, K.D., Tanaka, K., Tian, Q., Wanasinghe, D.N., Ariyawansa, H.A., Jayasiri, S.C., Boonmee, S., Camporesi, E., Hashimoto, A., Hirayama, K., Schumacher, R.K., Promputtha, I. & Liu, Z.-Y. (2015) Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. Nov. Fungal Diversity 74 (1): 199–266. https://doi.org/10.1007/s13225-015-0348-3
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Groenewald, J.Z., Cardinali, G., Houbraken, J., Boekhout, T., Crous, P.W., Robert, V. & Verkley, G.J.M. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92 (1): 135–154. https://doi.org/10.1016/j.simyco.2018.05.001
- Wanasinghe, D.N., Mortimer, P.E. & Xu, J. (2021) Insight into the Systematics of Microfungi Colonizing Dead Woody Twigs of Dodonaea viscosa in Honghe (China). Journal of Fungi 7 (3): 180. https://doi.org/10.3390/jof7030180
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In: PCR Protocols. Elsevier, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wimalasena, S.D.M.K., Wijayawardene, N.N., Dai, D.Q., Bamunuarachchige, T.C. & Jayalal, R.G.U. (2023) Morphological and molecular identification of Rhytidhysteron neorufulum found in aquatic environments of Anuradhapura district, Sri Lanka. In: Proceedings of the Young Scientists’ Conference on Multidisciplinary Research-2023. The Young Scientists Association of the National Institute of Fundamental Studies, Sri Lanka, pp. 35.
- Wong, T.K.F., Ly-Trong, N., Ren, H., Banos, H., Roger, A.J., Susko, E., Bielow, C., De Maio, N., Goldman, N. & Hahn, M.W. (2025) IQ-TREE 3: Phylogenomic Inference Software using Complex Evolutionary Models. [Submitted] https://doi.org/10.32942/X2P62N
- Xu, X.-L., Xiao, Q.-G., Yang, C.-L., Jeewon, R. & Liu, Y.-G. (2022) Multigene Phylogenetic Support for Novel Rhytidhysteron Speg. Species (Hysteriaceae) from Sichuan Province, China. Cryptogamie, Mycologie 43 (3): 63–79. https://doi.org/10.5252/cryptogamie-mycologie2022v43a3
- Zhao, L.-L., Zhang, L., Jiao, N., Li, M. & Zhang, Y. (2025) Branch blight and dieback of walnut (Juglans regia L.) caused by Rhytidhysteron juglandis sp. Nov. and its endophytic biocontrol agent Granulobasidium vellereum. Phytotaxa 705 (1): 19–34. https://doi.org/10.11646/phytotaxa.705.1.2
- Zhaxybayeva, O. & Gogarten, J.P. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 4. https://doi.org/10.1186/1471-2164-3-4