Abstract
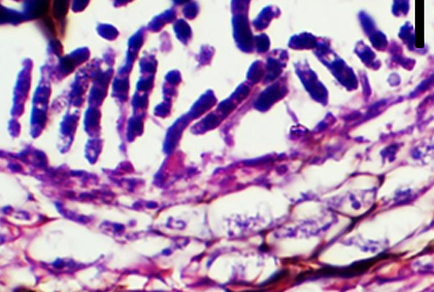
Vandijckomycella cardariae (Didymellaceae) is newly described and illustrated from the leaf spot of Cardaria draba. Phylogenetic analyses were based on the internal transcribed spacer regions 1 and 2 including the intervening 5.8S nuclear ribosomal DNA (ITS), partial β-tubulin (tub2), and RNA polymerase II second-largest subunit (rpb2). The strains of V. cardariae formed strongly supported clades, distinct from other previously known species in the phylogenetic tree. This new species is characterized by semi-immersed to immersed pycnidia having papillate or non-papillate ostioles, hyphal outgrowths, and sometimes with a short neck; pseudoparenchymatous cell walls; ampulliform to doliiform, phialidic conidiogenous cells; and cylindrical, aseptate, straight conidia, which are rounded at both ends. In addition, V. cardariae is easily distinguished from the two other species of Vandijckomycella by producing its cylindrical and longer conidia.
References
- Ahmadpour, S.A., Mehrabi-Koushki, M., Farokhinejad, R. & Asgari, B. (2022) New species of the family Didymellaceae in Iran. Mycological Progress 21 (2): 28. https://doi.org/10.1007/s11557-022-01800-5
- Ariyawansa, H.A., Tsai, I., Thambugala, K.M., Chuang, W.Y., Lin, S.R., Hozzein, W.N. & Cheewangkoon, R. (2020) Species diversity of Pleosporalean taxa associated with Camellia sinensis (L.) Kuntze in Taiwan. Scientific Reports 10 (1): 12762. https://doi.org/10.1038/s41598-020-69718-0
- Aveskamp, M.M., De Gruyter, J. & Crous, P.W. (2008) Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal diversity 31: 1–18.
- Aveskamp, M.M., de Gruyter, J., Woudenberg, J.H.C., Verkley, G.J.M. & Crous, P.W. (2010) Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in mycology 65 (1): 1–60. https://doi.org/10.3114/sim.2010.65.01
- Aveskamp, M.M., Woudenberg, J.H., De Gruyter, J., Turco, E., Groenewald, J.Z. & Crous, P.W. (2009) Development of taxon‐specific sequence characterized amplified region (SCAR) markers based on actin sequences and DNA amplification fingerprinting (DAF): a case study in the Phoma exigua species complex. Molecular Plant Pathology 10 (3): 403–414. https://doi.org/10.1111/j.1364-3703.2009.00540.x
- Chen, Q., Jiang, J.R., Zhang, G.Z., Cai, L. & Crous, P.W. (2015) Resolving the Phoma enigma. Studies in mycology 82: 137–217. https://doi.org/10.1016/j.simyco.2015.10.003
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature methods 9 (8): 772–772. https://doi.org/10.1038/nmeth.2109
- De Gruyter, J., Aveskamp, M.M., Woudenberg, J.H., Verkley, G.J. & Crous, P.W. (2009) Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycological research 113 (4): 508–519. https://doi.org/10.1016/j.mycres.2009.01.002
- Dudka, I.O., Heluta, V.P., Tykhonenko, Y.Y., Andrianova, T.V., Hayova, V.P., Prydiuk, M.P., Dzhagan, V.V. & Isikov, V.P. (2004) [Fungi of the Crimean Peninsula] (Translated from Russian). M.G. Kholodny Institute of Botany, National Academy of Sciences of Ukraine, 452 pp.
- Edler, D., Klein, J., Antonelli, A. & Silvestro, D. (2021) raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in ecology and evolution 12 (2): 373–377. https://doi.org/10.1111/2041-210X.13512
- Groenewald, M., Lombard, L., de Vries, M., Lopez, A.G., Smith, M. & Crous, P.W. (2018) Diversity of yeast species from Dutch garden soil and the description of six novel Ascomycetes. Federation of European Microbiological Societies Yeast Research 18 (7): foy076. https://doi.org/10.1093/femsyr/foy076
- Guo, S.Q., Norphanphoun, C., Hyde, K.D., Fu, S.M., Sun, J.E., Wang, X.C., Wu, J.J, Al-Otibi, F. & Wang, Y. (2025) Three novel species and a new record of Pleosporales (Didymellaceae, Roussoellaceae) from China. MycoKeys 113: 295–320. https://doi.org/10.3897/mycokeys.113.139934
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series 41 (41): 95–98.
- Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H.C., Sarma, V.V., Boonmee, S., Lücking, R., Bhat, D.J., Liu, N.G., Tennakoon, D.S., Pem, D., Karunarathna, A., Jiang, S.H., Jones, E.B.G., Phillips, A.J.L., Manawasinghe, I.S., Tibpromma, S., Jayasiri, S.C., Sandamali, D.S., Jayawardena, R.S., Wijayawardene, N.N., Ekanayaka, A.H., Jeewon, R., Lu, Y.Z., Dissanayake, A.J., Zeng, X.Y., Luo, Z.L., Tian, Q., Phukhamsakda, C., Thambugala, K.M., Dai, D.Q., Chethana, K.W.T., Samarakoon, M.C., Ertz, D., Bao, D.F., Doilom, M., Liu, J.K., Pérez-Ortega, S., Suija, A, Senwanna, C., Wijesinghe, S.N., Konta, S., Niranjan, M., Zhang, S.N., Ariyawansa, H.A., Jiang, H.B., Zhang, J.F., Norphanphoun, C., de, Silva, N.I., Thiyagaraja, V., Zhang, H., Bezerra, J.D.P., Miranda-González, R., Aptroot, A., Kashiwadani, H., Harishchandra, D., Sérusiaux, E., Aluthmuhandiram, J.V.S., Abeywickrama, P.D., Devadatha, B., Wu, H.X., Moon, K.H., Gueidan, C., Schumm, F., Bundhun, D., Mapook, A., Monkai, J., Chomnunti, P., Suetrong, S., Chaiwan, N., Dayarathne, M.C., Yang, J., Rathnayaka, A.R., Bhunjun, C.S., Xu, J., Zheng, J.S., Liu, G., Feng, Y. & Xie, N. (2020) Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105: 17–318. https://doi.org/10.1007/s13225-020-00462-6
- Hyde, K.D., Norphanphoun, C., Maharachchikumbura, S.S.N., Bhat, D.J., Jones, E.B.G., Bundhun, D., Chen, Y.J., Bao, D.F., Boonmee, S., Calabon, M.S., Chaiwan, N., Chethana, K.W.T., Dai, D.Q., Dayarathne, M.C., Devadatha, B., Dissanayake, A.J., Dissanayake, L.S., Doilom, M., Dong, W., Fan, X.L., Goonasekara, I.D., Hongsanan, S., Huang, S.K., Jayawardena, R.S., Jeewon, R., Karunarathna, A., Konta, S., Kumar, V., Lin, C.G., Liu, J.K., Liu, N.G., Luangsa-ard, J., Lumyong, S., Luo, Z.L., Marasinghe, D.S., McKenzie, E.H.C., Niego, A.G.T., Niranjan, M., Perera, R.H., Phukhamsakda, C., Rathnayaka, A.R., Samarakoon, M.C., Samarakoon, S.M.B.C, & Sarma, V.V., Senanayake, I.C., Shang, Q.J., Stadler, M., Tibpromma, S., Wanasinghe, D.N., Wei, D.P., Wijayawardene, N.N., Xiao, Y.P., Yang, J., Zeng, X.Y. & Zhang, S.N. (2020) Refined families of Sordariomycetes. Mycosphere 11 (1): 306–1060. https://doi.org/10.5943/mycosphere/11/1/7
- Hyde, K.D., Noorabadi, M.T., Thiyagaraja, V., He, M.Q., Johnston, P.R., Wijesinghe, S.N., Armand, A., Biketova, A.Y., Chethana, K.W.T., Erdoğdu, M., Ge, Z.W., Groenewald, J.Z., Hongsanan, S., Kušan, I., Leontyev, D.V., Li, D.W, Lin, C.G, Liu, N.G, Maharachchikumbura, S.S.N, Matočec, N, May, T.W, McKenzie, E.H.C, Mešić, A, Perera, R.H., Phukhamsakda, C., Piątek, M., Samarakoon, M.C., Selcuk, F., Senanayake, I.C., Tanney, J.B., Tian, Q., Vizzini, A., Wanasinghe, D.N., Wannasawang, N., Wijayawardene, N.N., Zhao, R.L., Abdel-Wahab, M.A., Abdollahzadeh, J., Abeywickrama, P.D., Abhinav, Absalan, S., Acharya, K., Afshari, N., Afshan, N.S., Afzalinia, S., Ahmadpour, S.A., Akulov, O., Alizadeh, A., Alizadeh, M., Al-Sadi, AM., Alves, A., Alves, V.C.S., Alves-Silva, G., Antonín, V., Aouali, S., Aptroot, A., Apurillo, C.C.S., Arias, R.M., Asgari, B., Asghari, R., Assis, D.M.A., Assyov, B., Atienza, V., Aumentado, H.D.R., Avasthi, S., Azevedo, E., Bakhshi, M., Bao, D.F., Baral, H.O., Barata, M., Barbosa, K.D., Barbosa, R.N., Barbosa, F.R., Baroncelli, R., Barreto, G.G., Baschien, C., Bennett, R.M., Bera, I., Bezerra, J.D.P., Bhunjun, C.S., Bianchinotti, M.V., Błaszkowski, J., Boekhout, T., Bonito, GM., Boonmee, S., Boonyuen, N., Bortnikov, FM., Bregant, C., Bundhun, D., Burgaud, G., Buyck, B., Caeiro, M.F., Cabarroi-Hernández, M., Cai, M., Feng, Cai, L., Calabon, M.S., Calaça, F.J.S., Callalli, M., Câmara, M.P.S., Cano-Lira, J., Cao, B., 5162, Carlavilla, J.R., Carvalho, A., Carvalho, T.G., Castañeda-Ruiz, R.F., Catania, M.D.V., Cazabonne, J., Cedeño-Sanchez, M., Chaharmiri-Dokhaharani, S., Chaiwan, N., Chakraborty, N., Cheewankoon, R., Chen, C., Chen, J., Chen, Q., Chen, Y.P., Chinaglia, S., Coelho-Nascimento, C.C., Coleine, C., CostaRezende, D.H., Cortés-Pérez, A., Crouch, J.A., Crous, P.W., Cruz, R.H.S.F., Czachura, P., Damm, U., Darmostuk, V., Daroodi, Z., Das, K., Das, K., Davoodian, N., Davydov, E.A., da Silva, G.A., da Silva, I.R., da Silva, R.M.F., da Silva, Santos, A.C., Dai, D.Q., Dai, Y.C., de Groot Michiel, D., De Kesel, A., De Lange, R., de Medeiros, E.V., de Souza, C.F.A., de Souza, F.A., dela Cruz, T.E.E., Decock, C., Delgado, G., Denchev, C.M., Denchev, T.T., Deng, Y.L., Dentinger, B.T.M., Devadatha, B., Dianese, J.C., Dima, B., Doilom, M., Dissanayake, A.J., Dissanayake, D.M.L.S, Dissanayake, L.S., Diniz, A.G., Dolatabadi, S., Dong, J.H., Dong, W., Dong, Z.Y., Drechsler-Santos, E.R., Druzhinina, I.S., Du, T.Y., Dubey, M.K., Dutta, A.K, Elliott, T.F., Elshahed, M.S., Egidi, E., Eisvand, P., Fan, L., Fan, X., Fan, X.L., Fedosova, A.G., Ferro, L.O., Fiuza, P.O., Flakus, A.W., Fonseca, E.O., Fryar, S.C., Gabaldón, T., Gajanayake, A.J, Gannibal, PB., Gao, F., GarcíaSánchez, D., García-Sandoval, R., Garrido-Benavent, I., Garzoli, L., Gasca-Pineda, J., Gautam, A.K., Gené, J., Ghobad-Nejhad, M., Ghosh, A., Giachini, A.J., Gibertoni, T.B., Gentekaki, E., Gmoshinskiy, V.I., GóesNeto, A., Gomdola, D., Gorjón, S.P., Goto, B.T., Granados-Montero, M.M., Griffith, G.W., Groenewald, M., Grossart, H-P., Gu, ZR., Gueidan, C., Gunarathne, A., Gunaseelan, S., Guo, S.L., Gusmão, L.F.P., Gutierrez, A.C., Guzmán-Dávalos, L., Haelewaters, D., Haituk, H., Halling, R.E., He, S.C., Heredia, G., HernándezRestrepo, M., Hosoya, T., Hoog, S.D., Horak, E, Hou, C.L., Houbraken, J., Htet, Z.H., Huang, S.K., Huang, W.J., Hurdeal, VG., Hustad, V.P., Inácio, C.A., Janik, P., Jayalal, R.G.U., Jayasiri, S.C., Jayawardena, R.S., Jeewon, R., Jerônimo, G.H., Jin, J., Jones, E.B.G., Joshi, Y., Jurjević, Ž., Justo, A., Kakishima, M., Kaliyaperumal, M., Kang, G.P., Kang, J.C., Karimi, O., Karunarathna, S.C., Karpov, S.A., Kezo, K., Khalid, A.N., Khan, M.K., Khuna, S., Khyaju, S., Kirchmair, M., Klawonn, I., Kraisitudomsook, N., Kukwa, M., Kularathnage, N.D., Kumar, S., Lachance, M.A., Lado, C., Latha, K.P.D., Lee, H.B., Leonardi, M., Lestari, A.S., Li, C., Li, H., Li, J., Li, Q., Li, Y., Li, Y.C., Li, Y.X., Liao, C.F., Lima, J.L.R., Lima, J.M.S, Lima, N.B., Lin, L., Linaldeddu, B.T., Linn, M.M., Liu, F., Liu, J.K., Liu, J.W., Liu, S., Liu, S.L., Liu, X.F., Liu, X.Y., Longcore, J.E., Luangharn, T., Luangsa-ard, J.J., Lu, L., Lu, Y.Z., Lumbsch, H.T., Luo, L., Luo, M., Luo, Z.L., Ma, J., Madagammana, A.D., Madhushan, A., Madrid, H., Magurno, F., Magyar, D., Mahadevakumar, S., Malosso, E., Malysh, J.M., Mamarabadi, M., Manawasinghe, I.S., Manfrino, R.G., Manimohan, P., Mao, N., Mapook, A., Marchese, P., Marasinghe, D.S., Mardones, M., Marin-Felix, Y., Masigol, H., Mehrabi, M., MehrabiKoushki, M., Meiras-Ottoni, A., de, Melo, R.F.R., Mendes-Alvarenga, R.L., Mendieta, S., Meng, Q.F., Menkis, A., Menolli, Jr, N., Mikšík, M., Miller, S.L., Moncada, B., Moncalvo, J.M., Monteiro, J.S., Monteiro, M., Mora-Montes, H.M., Moroz, E.L., Moura, J.C., Muhammad, U., Mukhopadhyay, S., Nagy, G.L., Najam, ul, Sehar, A., Najafiniya, M., Nanayakkara, C.M., Naseer, A., Nascimento, E.C.R., Nascimento, S.S., Neuhauser, S., Neves, M.A., Niazi, A.R., Nie, Yong., Nilsson, R.H., Nogueira, P.T.S., Novozhilov, Y.K., Noordeloos, M., Norphanphoun, C., Nuñez, Otaño, N., O’Donnell, R.P., Oehl, F., Oliveira, J.A., Oliveira,Junior, I., Oliveira, N.V.L., Oliveira, P.H.F., Orihara, T., Oset, M., Pang, K.L., Papp, V., Pathirana, L.S., Peintner, U., Pem, D., Pereira, O.L., Pérez-Moreno, J., Pérez-Ortega, S., Péter, G., Pires-Zottarelli, C.L.A., Phonemany, M., Phongeun, S., Pošta, A., Prazeres, J.F.S.A., Quan, Y., Quandt, C.A., Queiroz, M.B., Radek, R., Rahnama, K., Raj, K.N.A., Rajeshkumar, K.C., Rajwar Soumyadeep, Ralaiveloarisoa, A.B., Rämä, T., Ramírez-Cruz, V., Rambold, G., Rathnayaka, A.R., Raza, M., Ren, G.C., Rinaldi, A.C., Rivas-Ferreiro, M., Robledo, G.L., Ronikier, A., Rossi, W., Rusevska, K., Ryberg, M., Safi, A., Salimi, F., Salvador-Montoya, C.A., Samant, B., Samaradiwakara, N.P., Sánchez-Castro, I., Sandoval-Denis, M., Santiago, A.L.C.M.A., Santos, A.C.D.S., Santos, L.A., dos, Sarma, V.V., Sarwar, S., Savchenko, A., Savchenko, K., Saxena, R.K., Schoutteten, N., Selbmann, L., Ševčíková, H., Sharma, A., Shen, H.W., Shen, Y.M., Shu, Y.X., Silva, H.F., Silva-Filho, A.G.S, Silva, V.S.H., Simmons, D.R., Singh, R., Sir, E.B., Sohrabi, M., Souza, F.A., Souza-Motta, C.M., Sriindrasutdhi, V., Sruthi, O.P., Stadler, M., Stemler, J., Stephenson, S.L., Stoyneva-Gaertner, M.P., Strassert, J.F.H., Stryjak-Bogacka, M., Su, H., Sun, Y.R., Svantesson, S., Sysouphanthong, P., Takamatsu, S., Tan, T.H., Tanaka, K., Tang, C., Tang, X., Taylor, J.E., Taylor, P.W.J., Tennakoon, D.S., Thakshila, S.A.D., Thambugala, K.M., Thamodini, G.K., Thilanga, D., Thines, M., Tiago, P.V., Tian, X.G., Tian, W.H., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Tomšovský, M., Torruella, G., Tsurykau, A., Udayanga, D., Ulukapı, M., Untereiner, W.A., Usman, M., Uzunov, B.A., Vadthanarat, S., Valenzuela, R., Van den Wyngaert, S., Van Vooren, N., Velez, P, Verma, R.K., Vieira, L.C., Vieira, W.A.S., Vinzelj, J.M., Tang, A.M.C. & Walker, A.K., 5163 Wang, Q.M., Wang, Y., Wang, X.Y., Wang, Z.Y., Wannathes, N., Wartchow, F., Weerakoon, G., Wei, D.P., Wei, X., White, J.F., Wijesundara, D.S.A., Wisitrassameewong, K., Worobiec, G., Wu, H.X., Wu, N., Xiong, Y.R., Xu, B., Xu, J.P., Xu, R., Xu, R.F., Xu, R.J., Yadav, S., Yakovchenko, L.S., Yang, H.D., Yang, X., Yang, Y.H., Yang, Y., Yang, Y.Y, Yoshioka, R., Youssef Noha, H. & Yu, F.M., Yu, Z.F., Yuan, L.L., Yuan, Q., Zabin, D.A., Zamora, J.C., Zapata, C.V, Zare, R. (2024) The 2024 outline of fungi and fungus-like taxa. Mycosphere 15 (1): 5146–6239. https://doi.org/10.5943/mycosphere/15/1/25
- Hou, L., Hernández-Restrepo, M., Groenewald, J.Z., Cai, L. & Crous, P.W. (2020) Citizen science project reveals high diversity in Didymellaceae (Pleosporales, Dothideomycetes). MycoKeys 65: 49. https://doi.org/10.3897/mycokeys.65.47704
- Index Fungorum. (n.d.) Royal Botanic Gardens Kew. (accessed 10 January 2025)
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular biology and evolution 16 (12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Jayasiri, S.C., Hyde, K.D., Jones, E.B.G., Jeewon, R., Ariyawansa, H.A., Bhat, J.D., Camporesi, E. & Kang, J.C. (2017) Taxonomy and multigene phylogenetic evaluation of novel species in Boeremia and Epicoccum with new records of Ascochyta and Didymella (Didymellaceae). Mycosphere 8 (8): 1080–1101. https://doi.org/10.5943/mycosphere/8/8/9
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7 (11): 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Marin-Felix, Y., Groenewald, J.Z., Cai, L., Chen, Q., Marincowitz, S., Barnes, I., Bensch, K., Braun, U., Camporesi, E., Damm, U., de Beer, Z.W., Dissanayake, A., Edwards, J., Giraldo, A., Hernández-Restrepo, M., Hyde, K.D., Jayawardena, R.S., Lombard, L., Luangsa-ard, J., McTaggart, A.R., Rossman, A.Y., Sandoval-Denis, M., Shen, M., Shivas, R.G., Tan, Y.P., van der Linde, E.J. & Wingfield, M.J., Wood, A.R., Zhang, J.Q., Zhang, Y. & Crous, P.W. (2017) Genera of phytopathogenic fungi: GOPHY 1. Studies in mycology 86: 99–216. https://doi.org/10.1016/j.simyco.2017.04.002
- McKenzie, E.H.C. & Dingley, J.M. (1996) New plant disease records in New Zealand: miscellaneous fungal pathogens III. New Zealand journal of botany 34 (2): 263–272. https://doi.org/10.1080/0028825X.1996.10410690
- O’Donnell, K. & Cigelnik, E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungusfusariumare nonorthologous. Molecular phylogenetics and evolution 7 (1): 103–116. https://doi.org/10.1006/mpev.1996.0376
- Pem, D., Hyde, K.D., McKenzie, E.H.C., Hongsanan, S., Wanasinghe, D.N., Boonmee, S., Darmostuk, V., Bhat, J.D., Tian, Q., Htet, Z.H., Senanayake, I.C., Niranjan, M., Sarma, V.V., Doilom, M. & Dong, W. (2024) A comprehensive overview of genera in Dothideomycetes. Mycosphere 15 (1): 2175–4568. https://doi.org/10.5943/mycosphere/15/1/18
- Pem, D., Jeewon, R., Chethana, K.W.T., Hongsanan, S., Doilom, M., Suwannarach, N. & Hyde, K.D. (2021) Species concepts of Dothideomycetes: classification, phylogenetic inconsistencies and taxonomic standardization. Fungal Diversity 109 (1): 283–319. https://doi.org/10.1007/s13225-021-00485-7
- Priest, M.J. (2006) Fungi of Australia: Septoria. pp. vi + 1–259.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
- Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S. & Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11 (1): 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
- Sung, G.H., Sung, J.M., Hywel-Jones, N.L. & Spatafora, J.W. (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular phylogenetics and evolution 44 (3): 1204–1223. https://doi.org/10.1016/j.ympev.2007.03.011
- Tang, X., Jeewon, R., Lu, Y-Z., Alrefaei, A.F., Jayawardena, R.S., Xu, R-J., Ma, J., Chen, X-M. & Kang, J-C. (2023) Morphophylogenetic evidence reveals four new fungal species within Tetraplosphaeriaceae (Pleosporales, Ascomycota) from tropical and subtropical forest in China. MycoKeys 100: 171–204. https://doi.org/10.3897/mycokeys.100.113141
- Tang, X., Jeewon, R., Jayawardena, R.S., Gomdola, D., Lu, Y-Z., Xu, R-J., Alrefaei, A.F., Alotibi, F., Hyde, K.D. & Kang, J-C. (2024) Additions to the genus Kirschsteiniothelia (Dothideomycetes); Three novel species and a new host record, based on morphology and phylogeny. MycoKeys 110: 35–66. https://doi.org/10.3897/mycokeys.110.133450
- Wanasinghe, D.N., Jeewon, R., Peršoh, D., Jones, E.B.G., Camporesi, E., Bulgakov, T.S., Gafforov, Y.S. & Hyde, K.D. (2018) Taxonomic circumscription and phylogenetics of novel didymellaceous taxa with brown muriform spores. Studies in Fungi 3 (1): 152–175. https://doi.org/10.5943/sif/3/1/17
- White, T.J., Bruns, T., Lee, S.J. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18 (1): 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Woudenberg, J.H.C., Aveskamp, M.M., De Gruyter, J., Spiers, A.G. & Crous, P.W. (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia-Molecular Phylogeny and Evolution of Fungi 22 (1): 56–62. https://doi.org/10.3767/003158509X427808