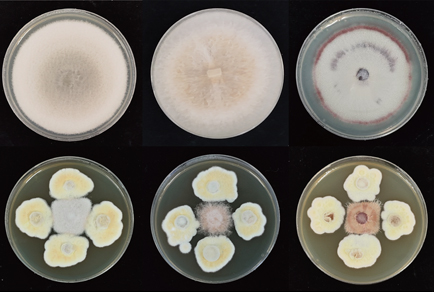
Abstract
Penicillium is a common, widespread and economically important genus. In an investigation of endophytic Penicillium species associated with trifoliate orange in China, a novel species in Penicillium section Exilicaulis, namely P. sinense was isolated from the leaves of Citrus trifoliata L. in Changsha, Hunan, China. It is different from other Penicillium species morphological comparisons and phylogenetic analyses. Multi-locus (CaM, ITS, RPB2 and TUB) phylogenetic analysis confirmed that P. sinense is distinct from other species. A synopsis of the morphological characters of the new species is provided. The biocontrol potential of P. sinense was measured by the confrontation test between P. sinense and common pathogenic fungi. The results of the study provided a strain resource for screening effective antagonistic Penicillium strains and biological control of fungal diseases.
References
- Arnold, A.E., Mejía, L.C., Kyllo, D., Rojas, E.I., Maynard, Z., Robbins, N. & Herre, E.A. (2003) Fungal endophytes limit pathogen damage in a tropical tree. Proceedings of the National Academy of Sciences of the United States of America 100: 15649–15654. https://doi.org/10.1073/pnas.2533483100
- Bhunjun, C.S., Chen, Y.J., Phukhamsakda, C., Boekhout, T., Groenewald, J.Z., Mckenzie, E.H.C., Francisco, E.C., Frisvad, J.C., Groenewald, M., Hurdeal, V.G., Luangsa-Ard, J., Perrone, G., Visagie, C.M., Bai, F.Y., Błaszkowski, J., Braun, U., de Souza, F.A., de Queiroz, M.B., Dutta, A.K., Gonkhom, D., Goto, B.T., Guarnaccia, V., Hagen, F., Houbraken, J., Lachance, M.A., Li, J.J., Luo, K.Y., Magurno, F., Mongkolsamrit, S., Robert, V., Roy, N., Tibpromma, S., Wanasinghe, D.N., Wang, D.Q., Wei, D.P., Zhao, C.L., Aiphuk, W., Ajayi-Oyetunde, O., Arantes, T.D., Araujo, J.C., Begerow, D., Bakhshi, M., Barbosa, R.N., Behrens, F.H., Bensch, K., Bezerra, J.D.P., Bilaski, P., Bradley, C.A., Bubner, B., Burgess, T.I., Buyck, B., Adež, N., Cai, L., Calaça, F.J.S., Campbell, L.J., Chaverri, P., Chen, Y.Y., Chethana, K.W.T., Coetzee, B., Costa, M.M., Chen, Q., Custódio, F.A., Dai, Y.C., Damm, U., Santiago, A.L.C.M.A., De Angelini, R.M. Miccolis, Dijksterhuis, J., Dissanayake, A.J., Doilom, M., Dong, W., álvarez-Duarte, E., Fischer, M., Gajanayake, A.J., Gené, J., Gomdola, D., Gomes, A.A.M., Hausner, G., He, M.Q., Hou, L., Iturrieta-González, I., Jami, F., Jankowiak, R., Jayawardena, R.S., Kandemir, H., Kiss, L., Kobmoo, N., Kowalski, T., Landi, L., Lin, C.G., Liu, J.K., Liu, X.B., Loizides, M., Luangharn, T., Maharachchikumbura, S.S.N., Mkhwanazi, G.J. Makhathini, Manawasinghe, I.S., Marin-Felix, Y., Mctaggart, A.R., Moreau, P.A., Morozova, O.V., Mostert, L., Osiewacz, H.D., Pem, D., Phookamsak, R., Pollastro, S., Pordel, A., Poyntner, C., Phillips, A.J.L., Phonemany, M., Promputtha, I., Rathnayaka, A.R., Rodrigues, A.M., Romanazzi, G., Rothmann, L., Salgado-Salazar, C., Sandoval-Denis, M., Saupe, S.J., Scholler, M., Scott, P., Shivas, R.G., Silar, P., Silva-Filho, A.G.S., Souza-Motta, C.M., Spies, C.F.J., Stchigel, A.M., Sterflinger, K., Summerbell, R.C., Svetasheva, T.Y., Takamatsu, S., Theelen, B., Theodoro, R.C., Thines, M., Thongklang, N., Torres, R., Turchetti, B., Van Den Brule, T., Wang, X.W., Wartchow, F., Welti, S., Wijesinghe, S.N., Wu, F., Xu, R., Yang, Z.L., Yilmaz, N., Yurkov, A., Zhao, L., Zhao, R.L., Zhou, N., Hyde, K.D. & Crous, P.W. (2024) What are the 100 most cited fungal genera? Studies in Mycology 108: 1–411. https://doi.org/10.3114/sim.2024.108.01
- Cai, F., Yang, C., Ma, T., Osei, R., Jin, M., Zhang, C. & Wang, Y. (2024) An endophytic Paenibacillus polymyxa hg18 and its biocontrol potential against Fusarium oxysporum f. sp. cucumerinum. Biological Control 188: 105380.
- Du, C., Lin, S.K., Koutinas, A., Wang, R., Dorado, P. & Webb, C. (2008) A wheat biorefining strategy based on solid-state fermentation for fermentative production of succinic acid. Bioresource Technol 99: 8310–8315. https://doi.org/10.1016/j.biortech.2008.03.019
- Guo, L.D., Hyde, K.D. & Liew, E.C.Y. (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol 147: 617–630. https://doi.org/10.1046/j.1469-8137.2000.00716.x
- Houbraken, J. & Samson, R.A. (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70: 1–51. https://doi.org/10.3114/sim.2011.70.01
- Houbraken, J., Frisvad, J.C. & Samson, R.A. (2011) Taxonomy of Penicillium section Citrina. Stud Mycol 70: 53–138. https://doi.org/10.3114/sim.2011.70.02
- Houbraken, J., Kocsubé, S., Visagie, C.M., Yilmaz, N., Wang, X.-C., Meijer, M., Kraak, B., Hubka, V., Bensch, K., Samson, R.A. & Frisvad, J.C. (2020) Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169.
- Houbraken, J., Visagie, C.M., Meijer, M., Frisvad, J.C., Busby, P.E., Pitt, J.I., Seifert, K.A., Louis-Seize, G., Demirel, R., Yilmaz, N., Jacobs, K., Christensen, M. & Samson, R.A.A. (2014) taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud Mycol 78: 373–451. https://doi.org/10.1016/j.simyco.2014.09.002
- Huang, L.Q., Niu, Y.C., Su, L., Deng, H. & Lyu, H. (2020) The potential of endophytic fungi isolated from cucurbit plants for biocontrol of soilborne fungal diseases of cucumber. Microbiological Research 231: 126369.
- Hyde, K.D., Noorabadi, M.T., Thiyagaraja, V., He, M.Q., Johnston, P.R., Wijesinghe, S.N., Armand, A., Biketova, A.Y., Chethana, K.W.T., Erdoğdu, M., Ge, Z.W., Groenewald, J.Z., Hongsanan, S., Kušan, I., Leontyev, D.V., Li, D.W., Lin, C.G., Liu, N.G., Maharachchikumbura, S.S.N., Matočec, N., May, T.W., McKenzie, E.H.C., Mešić, A., Perera, R.H., Phukhamsakda, C., Piątek, M., Samarakoon, M.C., Selcuk, F., Senanayake, I.C., Tanney, J.B., Tian, Q., Vizzini, A., Wanasinghe, D.N., Wannasawang, N., Wijayawardene, N., Zhao, R.L., Abdel-Wahab, M.A., Abdollahzadeh, J., Abeywickrama, P.D., Abhinav, Absalan, S., Acharya, K., Afshari, N., Afshan,. NS, Afzalinia, S., Ahmadpour, S.A., Akulov, O., Alizadeh, A., Alizadeh, M., Al-Sadi, A.M., Alves, A., Alves, V.C.S., Alves-Silva, G., Antonín, V., Aouali, S., Aptroot, A., Apurillo, C.C.S., Arias, R.M., Asgari, B., Asghari, R., Assis, D.M.A., Assyov, B., Atienza, V., Aumentado, H.D.R., Avasthi, S., Azevedo, E., Bakhshi, M., Bao, D.F., Baral, H.O., Barata, M., Barbosa, K.D., Barbosa, R.N., Barbosa, F.R., Baroncelli, R., Barreto, G.G., Baschien, C., Bennett, R.M., Bera, I., Bezerra, J.D.P., Bhunjun, C.S., Bianchinotti, M.V., Błaszkowski, J., Boekhout, T., Bonito, G.M., Boonmee, S., Boonyuen, N., Bortnikov, F.M., Bregant, C., Bundhun, D., Burgaud, G., Buyck, B., Caeiro, M.F., Cabarroi-Hernández, M., Cai, M.F., Cai, L., Calabon, M.S., Calaça, F.J.S., Callalli, M., Câmara, M.P.S., Cano-Lira, J., Cao, B., Carlavilla, J.R., Carvalho, A., Carvalho, T.G., Castañeda-Ruiz, R.F., Catania, M.D.V., Cazabonne, J., Cedeño-Sanchez, M., Chaharmiri-Dokhaharani, S., Chaiwan, N., Chakraborty, N., Cheewankoon, R., Chen, C., Chen, J., Chen, Q., Chen, Y.P., Chinaglia, S., Coelho-Nascimento, C.C., Coleine, C., CostaRezende, D.H., Cortés-Pérez, A., Crouch, J.A., Crous, P.W., Cruz, R.H.S.F., Czachura, P., Damm, U., Darmostuk, V., Daroodi, Z., Das, K., Davoodian, N., Davydov, E.A., da Silva, G.A., da Silva, I.R., da Silva, R.M.F., da Silva, S.A.C., Dai, D.Q., Dai, Y.C., de Groot, M.D., De Kesel, A., De Lange, R., de Medeiros, E.V., de Souza, C.F.A., de Souza, F.A., dela Cruz, T.E.E., Decock, C., Delgado, G., Denchev, C.M., Denchev, T.T., Deng, Y.L., Dentinger, B.T.M., Devadatha, B., Dianese, J.C., Dima, B., Doilom, M., Dissanayake, A.J., Dissanayake, D.M.L.S., Dissanayake, L.S., Diniz, A.G., Dolatabadi, S., Dong, J.H., Dong, W., Dong, Z.Y., Drechsler-Santos, E.R., Druzhinina, I.S., Du, T.Y., Dubey, M.K., Dutta, A.K., Elliott, T.F., Elshahed, M.S., Egidi, E., Eisvand, P., Fan, L., Fan, X., Fan, X.L., Fedosova, A.G., Ferro, L.O., Fiuza, P.O., Flakus, A.W., Fonseca, E.O., Fryar, S.C., Gabaldón, T., Gajanayake, A.J., Gannibal, P.B., Gao, F., GarcíaSánchez, D., García-Sandoval, R., Garrido-Benavent, I., Garzoli, L., Gasca-Pineda, J., Gautam, A.K., Gené, J., Ghobad-Nejhad, M., Ghosh, A., Giachini, A.J., Gibertoni, T.B., Gentekaki, E., Gmoshinskiy, V.I., GóesNeto, A., Gomdola, D., Gorjón, S.P., Goto, B.T., Granados-Montero, M.M., Griffith, G.W., Groenewald, M., Grossart, H.-P., Gu, Z.R., Gueidan, C., Gunarathne, A., Gunaseelan, S., Guo, S.L., Gusmão, L.F.P., Gutierrez, A.C., Guzmán-Dávalos, L., Haelewaters, D., Haituk, H., Halling, R.E., He, S.C., Heredia, G., HernándezRestrepo, M., Hosoya, T., Hoog, S.D., Horak, E., Hou, C.L., Houbraken, J., Htet, Z.H., Huang, S.K., Huang, W.J., Hurdeal, V.G., Hustad, V.P., Inácio, C.A., Janik, P., Jayalal, R.G.U., Jayasiri, S.C., Jayawardena, R.S., Jeewon, R., Jerônimo, G.H., Jin, J., Jones, E.B.G., Joshi, Y., Jurjević, Ž., Justo, A., Kakishima, M., Kaliyaperumal, M., Kang, G.P., Kang, J.C., Karimi, O., Karunarathna, S.C., Karpov, S.A., Kezo, K., Khalid, A.N., Khan, M.K., Khuna, S., Khyaju, S., Kirchmair, M., Klawonn, I., Kraisitudomsook, N., Kukwa, M., Kularathnage, N.D., Kumar, S., Lachance, M.A., Lado, C., Latha, K.P.D., Lee, H.B., Leonardi, M., Lestari, A.S., Li, C., Li, H., Li, J., Li, Q., Li, Y., Li, Y.C., Li, Y.X., Liao, C.F., Lima, J.L.R., Lima, J.M.S., Lima, N.B., Lin, L., Linaldeddu, B.T., Linn, M.M., Liu, F., Liu, J.K., Liu, J.W., Liu, S., Liu, S.L., Liu, X.F., Liu, X.Y., Longcore, J.E., Luangharn, T., Luangsaard, J.J., Lu, L., Lu, Y.Z., Lumbsch, H.T., Luo, L., Luo, M., Luo, Z.L., Ma, J., Madagammana, A.D., Madhushan, A., Madrid, H., Magurno, F., Magyar, D., Mahadevakumar, S., Malosso, E., Malysh, J.M., Mamarabadi, M., Manawasinghe, I.S., Manfrino, R.G., Manimohan, P., Mao, N., Mapook, A., Marchese, P., Marasinghe, D.S., Mardones, M., Marin-Felix, Y., Masigol, H., Mehrabi, M., MehrabiKoushki, M., Meiras-Ottoni, A., de Melo, R.F.R., Mendes-Alvarenga, R.L., Mendieta, S., Meng, Q.F., Menkis, A., Menolli, J.N., Mikšík, M., Miller, S.L., Moncada, B., Moncalvo, J.M., Monteiro, J.S., Monteiro, M., Mora-Montes, H.M., Moroz, E.L., Moura, J.C., Muhammad, U., Mukhopadhyay, S., Nagy, G.L., Najamul, S.A., Najafiniya, M., Nanayakkara, C.M., Naseer, A., Nascimento, E.C.R., Nascimento, S.S., Neuhauser, S., Neves, M.A., Niazi, A.R., Nie, Yong., Nilsson, R.H., Nogueira, P.T.S., Novozhilov, Y.K., Noordeloos, M., Norphanphoun, C., Nuñez, O.N., O’Donnell, R.P., Oehl, F., Oliveira, J.A., Oliveira, J.I., Oliveira, N.V.L., Oliveira, P.H.F., Orihara, T., Oset, M., Pang, K.L., Papp, V., Pathirana, L.S., Peintner, U., Pem, D., Pereira, O.L., Pérez-Moreno, J., Pérez-Ortega, S., Péter, G., Pires-Zottarelli, C.L.A., Phonemany, M., Phongeun, S., Pošta, A., Prazeres, J.F.S.A., Quan, Y., Quandt, C.A., Queiroz, M.B., Radek, R., Rahnama, K., Raj, K.N.A., Rajeshkumar, K.C., Rajwar, S., Ralaiveloarisoa, A.B., Rämä, T., Ramírez-Cruz, V., Rambold, G., Rathnayaka, A.R., Raza, M., Ren, G.C., Rinaldi, A.C., Rivas-Ferreiro, M., Robledo, G.L., Ronikier, A., Rossi, W., Rusevska, K., Ryberg, M., Safi, A., Salimi, F., Salvador-Montoya, C.A., Samant, B., Samaradiwakara, N.P., Sánchez-Castro, I., Sandoval-Denis, M., Santiago, A.L.C.M.A., Santos, A.C.D.S., Santos, L.A., Sarma, V.V., Sarwar, S., Savchenko, A., Savchenko, K., Saxena, R.K., Schoutteten, N., Selbmann, L., Ševčíková, H., Sharma, A., Shen, H.W., Shen, Y.M., Shu, Y.X., Silva, H.F., Silva-Filho, A.G.S., Silva, V.S.H., Simmons, D.R., Singh, R., Sir, E.B., Sohrabi, M., Souza, F.A., Souza-Motta, C.M., Sriindrasutdhi, V., Sruthi, O.P., Stadler, M., Stemler, J., Stephenson, S.L., Stoyneva-Gaertner, M.P., Strassert, J.F.H., Stryjak-Bogacka, M., Su, H., Sun, Y.R., Svantesson, S., Sysouphanthong, P., Takamatsu, S., Tan, T.H., Tanaka, K., Tang, C., Tang, X., Taylor, J.E., Taylor, P.W.J, Tennakoon, D.S., Thakshila, S.A.D., Thambugala, K.M., Thamodini, G.K., Thilanga, D., Thines, M., Tiago, P.V., Tian, X.G., Tian, W.H., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Tomšovský, M., Torruella, G., Tsurykau, A., Udayanga, D., Ulukapı, M., Untereiner, W.A., Usman, M., Uzunov, B.A., Vadthanarat, S., Valenzuela, R., Van den Wyngaert, S., Van Vooren, N., Velez, P., Verma, R.K., Vieira, L.C., Vieira, W.A.S., Vinzelj, J.M., Tang, A.M.C., Walker, A., Walker, A.K., Wang, Q.M., Wang, Y., Wang, X.Y., Wang, Z.Y., Wannathes, N., Wartchow, F., Weerakoon, G., Wei, D.P., Wei, X., White, J.F., Wijesundara, D.S.A., Wisitrassameewong, K., Worobiec, G., Wu, H.X., Wu, N., Xiong, Y.R., Xu, B., Xu, J.P., Xu, R., Xu, R.F., Xu, R.J., Yadav, S., Yakovchenko, L.S., Yang, H.D., Yang, X., Yang, Y.H., Yang, Y., Yang, Y.Y., Yoshioka, R., Youssef, N.H., Yu, F.M., Yu, Z.F., Yuan, L.L., Yuan, Q., Zabin, D.A., Zamora, J.C., Zapata, C.V., Zare, R., Zeng, M., Zeng, X.Y., Zhang, J.F., Zhang, J.Y., Zhang, S., Zhang, X.C., Zhao, C.L., Zhao, H., Zhao, Q., Zhao, H., Zhao, H.J., Zhou, H.M., Zhu, X.Y., Zmitrovich, I.V., Zucconi, L. & Zvyagina, E. (2024) The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 15 (1): 5146–6239. https://doi.org/10.5943/mycosphere/15/1/25
- Kremer, A.M., Pal, T.M., de Monchy, J.G., Kauffman, H.F. & de Vries, K. (1989) Precipitating antibodies and positive skin tests in workers exposed to airborne antigens from a contaminated humidification system. Int Arch Occup Environ Health 61: 547–553. https://doi.org/10.1007/BF00683125
- Lu, Q.H., Wang, Y.C. & Li, Nana., Ni, D.J., Yang, Y.J. & Wang, X.C. (2018) Differences in the characteristics and pathogenicity of Colletotrichum camelliae and C. fructicola isolated from the tea plant [Camellia sinensis (L.) O. Kuntze]. Frontiers in Microbiology 9: 3060.
- Lyratzopoulos, G., Ellis, M., Nerringer, R. & Denning, D.W. (2002) Invasive Infection due to Penicillium species other than P. marneffei. Journal of Infection 45: 184–195. https://doi.org/10.1053/jinf.2002.1056
- McMullin, D.R., Nsiama, T.K. & Miller, J.D. (2014) Secondary metabolites from Penicillium corylophilum isolated from damp buildings. Mycologia 106: 621–628. https://doi.org/10.3852/13–265
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, USA, pp. 1–8.
- Miyake, I. (1940) Studies on toxin production by a saprophyte growing on stored rice. Report of the Rice Utilization Laboratories, Hôkoku 1: 1–30.
- Ohnishi, T., Yamada, G., Tanaka, H., Nakajima, K., Tanaka, S., Morita-Ichimura, S., Takahashi, R., Sato, M., Shibusa, T. & Abe, S. (2002) A case of chronic hypersensitivity pneumonia with elevation of serum SP-D and KL-6. Nihon Kokyuki Gakkai Zasshi 40: 66–70.
- Peterson, S.W., Corneli, S., Hjelle, J., Miller-Hjelle, M., Nowak, D. & Bonneau, P. (1999) Penicillium pimiteouiense: A new species isolated from polycystic kidney cell cultures. Mycologia 91: 269–277.
- Peterson, S.W., Orchard, S.S. & Menon, S. (2011) Penicillium menonorum, a new species related to P. pimiteouiense. IMA Fungus 2: 121–125. https://doi.org/10.5598/imafungus.2011.02.02.02
- Posada, D. & Crandall, K.A. (1998) MODELTEST: Testing the model of DNA substitution. Bioinformatics 14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817
- Redman, R.S., Sheehan, K.B., Stout, R.G., Rodriguez, R.J. & Henson, J.M. (2002) Thermotolerance generated by plant/fungal symbiosis. Science 298: 1581. https://doi.org/10.1126/science.1072191
- Rivera, K.G. & Seifert, K.A. (2011) A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Studies in Mycology 70: 139–158. https://doi.org/10.3114/sim.2011.70.03
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Rosa, C.A., Keller, K.M., Oliveira, A.A., Almeida, T.X., Keller, L.A., Marassi, A.C., Kruger, C.D., Deveza, M.V., Monteiro, B.S., Nunes, L.M., Astoreca, A., Cavaglieri, L.R., Direito, G.M., Eifert, E.C., Lima, T.A., Modernell, K.G., Nunes, F.I., Garcia, A.M., Luz, M.S. & Oliveira, D.C. (2010) Production of citreoviridin by Penicillium citreonigrum strains associated with rice consumption and beriberi cases in the Maranhão State, Brazil. Food Addit Contam 27: 24–248. https://doi.org/10.1080/19440040903289712
- Schulz, B., Guske, S., Dammann, U. & Boyle, C. (1998) Endophyte-host interactions. II. Defining symbiosis of the endophyte-host interaction. Symbiosis 25: 213–227.
- Serra, R., Peterson, S. & Venâncio, A. (2008) Multilocus sequence identification of Penicillium species in cork bark during plank preparation for the manufacture of stoppers. Research in Microbiology 159: 178–186. https://doi.org/10.1016/j.resmic.2007.12.009
- Stamatakis, A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446
- Su, L., Deng, H. & Niu, Y.C. (2016) Phialemoniopsis endophytica sp. nov., a new species of endophytic fungi from Luffa cylindrica in Henan, China. Mycol Prog 15: 48. https://doi.org/10.1007/s11557-016-1189-5
- Su, L., Deng, H. & Niu, Y.C. (2017) Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycological Progress 16 (8): 823–829.
- Su, L., Hao, Y., Xiang, M.C., Cai, L., Guo, L.Y. & Liu, X.Z. (2015) Rupestriomyces and Spissiomyces, two new genera of rock-inhabiting fungi from China. Mycologia 107 (4): 831.
- Su, L. & Niu, Y.C. (2018) Multi-locus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species, T. cucurbitiradicus and T. endophyticus. Mycologia 110 (2): 375–386.
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. & Kumar, S. (2011) MEGA 5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. https://doi.org/10.1093/molbev/msr121
- Taylor, J.W., Jacobson, D.J., Kroken, S., Kasuga, T., Geiser, D.M., Hibbett, D.S. & Fisher, M.C. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. https://doi.org/10.1006/fgbi.2000.1228
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. & Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. https://doi.org/10.1093/nar/25.24.4876
- Unoura, K., Miyazaki, Y., Sumi, Y., Tamaoka, M., Sugita, T. & Inase, N. (2011) Identification of fungal DNA in BALF from patients with homerelated hypersensitivity pneumonitis. Respiratory Medicine 105: 1696–1703. https://doi.org/10.1016/j.rmed.2011.07.009
- Visagie, C.M., Houbraken, J., Frisvad, J.C., Hong, S.B., Klaassen, C.H., Perrone, G., Seifert, K.A., Varga, J., Yaguchi, T. & Samson, R.A. (2014) Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. https://doi.org/10.1016/j.simyco.2014.09.001
- Visagie, C.M., Houbraken, J., Rodriques, C., Silva Pereira, C., Dijksterhuis, J., Seifert, K.A., Jacobs, K. & Samson, R.A. (2013) Five new Penicillium species in section Sclerotiora: a tribute to the dutch royal family. Persoonia 31: 42–62. https://doi.org/10.3767/003158513X667410
- Visagie, C.M., Seifert, K.A., Houbraken, J., Samson, R.A. & Jacobs, K. (2016) A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 7: 75–117. https://doi.org/10.5598/imafungus.2016.07.01.06
- Wei, X.C., Wang, Q., Shi, B., Xie, X.W., Shi, Y.X., Chai, A.L., Li, B.J. & Li, L. (2024) Isolation and Identification of Burkholderia cenocepacia YM12 and Its Control Effect on Cucumber Soft Rot Disease. Chinese Journal of Biological Control 40: 1088–1098.
- Yoshida, K., Hiraoka, T., Ando, M., Uchida, K. & Mohsenin, V. (1992) Penicillium decumbens: a new cause of fungus ball. Chest 101: 1152–1153. https://doi.org/10.1378/chest.101.4.1152
- Zhang, Y.-H., Du, H.-F., Song, J., Zhang, J., Chen, F., An, H., Cao, F. & Luo, D.-Q. (2025) Effect and mechanism of patchouli alcohol for the management of postharvest green pepper fruit rot caused by Alternaria alternata. Journal of Agricultural and Food Chemistry 73 (9): 5274–5288.
- Zhao, X.B., Ni, Y.X., Liu, X.T., Zhao, H., Yan, W.Q., He, B.P. & Liu, H.Y. (2023) Analysis of inhibitiory active substances of Penicillium bilaiae strain 47M-1 based on non-targeted metabolomics. Chinese Journal of Biological Control 39: 1156–1171.