Abstract
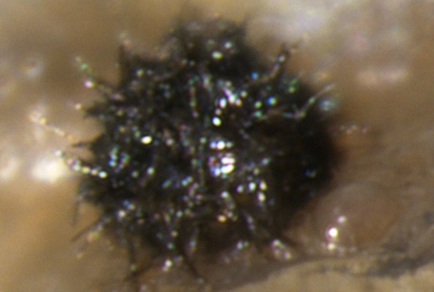
During a survey of soil fungi in rocky desertification regions of Yunnan Province, China, a novel species, Pyrenochaetopsis huizeanum, was isolated from nutrient-poor soils. Phylogenetic analyses using combined ITS, LSU, rpb2 and tub2 gene sequences clearly positioned the new species within the genus Pyrenochaetopsis. The novel taxon formed a distinct monophyletic lineage closely related to a group of six known species viz. P. kuksensis, P. musae, P. oryzicola, P. paucisetosa, P. botulispora and P. tabarestanensis. Morphologically, Pyrenochaetopsis huizeanum differs from P. musae, P. oryzicola and P. botulispora in having setae and larger pycnidia, from P. paucisetosa by having narrower conidia, from P. kuksensis by producing larger conidia, and from P. tabarestanensis by lacking chlamydospore. Here, we provide a detailed description, illustrations, and phylogenetic placement of the newly proposed species, Pyrenochaetopsis huizeanum.
References
- Absalan, S., Armand, A., Jayawardena, R.S., McKenzie, E.H.C. & Lumyong, S. (2024) Diversity of Pleosporalean Fungi Isolated from Rice (Oryza sativa L.) in Northern Thailand and Descriptions of Five New Species. Journal of Fungi 10 (11). https://doi.org/10.3390/jof10110763
- Carvalho, J.L., Lima, J.M., Barbier, E., Bernard, E., Bezerra, J.D. & Souza-Motta, C.M. (2022) Ticket to ride: fungi from bat ectoparasites in a tropical cave and the description of two new species. Brazilian Journal of Microbiology 53 (4): 2077–2091. https://doi.org/10.1007/s42770-022-00841-y
- Crous, P.W., Cowan, D.A., Maggs-Kölling, G., Yilmaz, N., Larsson, E., Angelini, C., Brandrud, T.E., Dearnaley, J.D.W., Dima, B., Dovana, F., Fechner, N., Garcia, D., Gené, J., Halling, R.E., Houbraken, J., Leonard, P., Luangsa-ard, J.J., Noisripoom, W., Rea-Ireland, A.E., Ševčíková, H., Smyth, C.W., Vizzini, A., Adam, J.D., Adams, G.C., Alexandrova, A.V., Alizadeh, A., Álvarez Duarte, E., Andjic, V., Antonín, V., Arenas, F., Assabgui, R., Ballarà, J., Banwell, A., Berraf-Tebbal, A., Bhatt, V.K., Bonito, G., Botha, W., Burgess, T.I., Caboň, M., Calvert, J., Carvalhais, L.C., Courtecuisse, R., Cullington, P., Davoodian, N., Decock, C.A., Dimitrov, R., Di Piazza, S., Drenth, A., Dumez, S., Eichmeier, A., Etayo, J., Fernandez, I., Fiard, J-P, Fournier, J., Fuentes-Aponte, S., Ghanbary, M.A.T., Ghorbani, G., Giraldo, A., Glushakova, A.M., Gouliamova, D.E., Guarro, J., Halleen, F., Hampe, F., Hernández-Restrepo, M., Iturrieta-González, I., Jeppson, M., Kachalkin, A.V., Karimi, O., Khalid, A.N., Khonsanit, A., Kim, J.I., Kim, K., Kiran, M., Krisai-Greilhuber, I., Kučera, V., Kušan, I., Langenhoven, S.D., Lebel, T., Lebeuf, R., Liimatainen, K., Linde, C., Lindner, D.L., Lombard, L., Mahamedi, A.E., Matočec, N., Maxwell, A., May, T.W., McTaggart, A.R., Meijer, M., Mešić, A., Mileto, A.J., Miller, A.N., Molia, A., Mongkolsamrit, S., Muñoz Cortés, C., Muñoz-Mohedano, J., Morte, A., Morozova, O.V., Mostert, L., Mostowfizadeh-Ghalamfarsa, R., Nagy, L.G., Navarro-Ródenas, A., Örstadius, L., Overton, B.E., Papp, V., Para, R., Peintner, U., Pham, T.H.G., Pordel, A., Pošta, A., Rodríguez, A., Romberg, M., Sandoval-Denis, M., Seifert, K.A., Semwal, K.C., Sewall, B.J., Shivas, R.G., Slovák, M., Smith, K., Spetik, M., Spies, C.F.J., Syme, K., Tasanathai, K., Thorn, R.G., Tkalčec, Z., Tomashevskaya, M.A., Torres-Garcia, D., Ullah, Z., Visagie, C.M., Voitk, A. & Groenewald, J.Z. (2020) Fungal Planet description sheets: 1112–1181. Persoonia - Molecular Phylogeny and Evolution of Fungi 45 (1): 251–409. https://doi.org/10.3767/persoonia.2020.45.10
- de Gruyter, J. & Boerema, G.H. (2002) Contributions towards a monograph of Phoma (Coelomycetes) VIII. Section Paraphoma: Taxa with setose pycnidia. Persoonia-Molecular Phylogeny and Evolution of Fungi 541–561.
- de Gruyter, J., Aveskamp, M.M., Woudenberg, J.H., Verkley, G.J. & Crous, P.W. (2009) Molecular phylogeny of Phoma and allied anamorph genera: towards a reclassification of the Phoma complex. Mycological research 113 (4): 508–519. https://doi.org/10.1016/j.mycres.2009.01.002
- de Gruyter, J., Woudenberg, J.H.C., Aveskamp, M.M., Verkley, G.J.M., Groenewald, J.Z. & ous, P.W. (2010) Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 102: 1066–1081. https://doi.org/10.3852/09–240
- Gams, W., Hoekstra, E.S. & Aptroot, A. (1998) CBS Course of Mycology, 4th ed. Centraalbureau voor Schimmelcultures, Baarn.
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford,England) 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Hyde, K.D., Tennakoon, D.S., Jeewon, R., Bhat, D.J., Maharachchikumbura, S.S., Rossi, W., Leonardi, M., Lee, H.B., Mun, H.Y., Houbraken, J., Nguyen, T.T.T., Jeon, S.J., Frisvad, J.C., Wangasinghe, D.N., Lücking, R., Aptroot, A., Cáceres, M.E.S., Karunarathna, S.C., Hongsanan, S., Phookamsak, R., de Silva, N.L., Thambugala, K.M., Jayawardena, R.S., Senanayake, I.C., Boonmee, S., Chen, J., Luo, Z.L., Phukhamsakda, C., Pereira, O.L., Abreu, V.P., Rosado, A.M.S., Sotão, H.M.P., Xavier, W.K.S., Bezerra, J.D.P., de Oliveira, T.G.L., de Souza-Motta, C.M., Oliane Magalhães, M.C., Bundhun, D., Harishchandra, D., Manawasinghe, I.S., Dong, W., Zhang, S.N., Bao, D.F., Samarakoon, M.C., Pem, D., Karunarathna, A., Lin, C.G., Yang, J., Perera, R.H., Kumar, V., Huang, S.K., Dayarathne, M.C., Ekanayaka, A.H., Jayasiri, S.C., Xiao, Y., Konta, S., Niskanen, T., Liimatainen, K., Dai, Y.C., Ji, X.H., Tian, X.M., Mešić, A., Singh, S.K., Phutthacharoen, K., Cai, L., Sorvongxay, T., Thiyagaraja, V., Norphanphoun, C., Chaiwan, N., Lu, Y.Z., Jiang, H.B., Zhang, J.F., Abeywickrama, P.D., Aluthmuhandiram, J.V.S., Brahmanage, R.S., Zeng, M., Chethana, T., Wei, D., Réblová, M., Fournier, J., Nekvindová, J., Barbosa, R.N., dos Santos, J.E.F., de Oliveira, N.T., Li, G.J., Ertz, D., Shang, Q.J., Phillips, A.J.L., Kuo, C.H., Camporesi, E., Bulgakov, T.S., Lumyong, S., Jones, E.B.G., Chomnunti, P., Gentekaki, E., Bungartz, F., Zeng, X.Y., Fryar, S., Tkalčec, Z., Liang, J., Li, G.S., Wen, T.C., Singh, P.N., Gafforov, Y., Promputtha, I., Yasanthika, E., Goonasekara, I.D., Zhao, R.L., Zhao, Q., Kirk, P.M., Liu, J.K. & Yan, J.Y., Mortimer, P.E., Xu, J.C. & Doilom, M.K. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal diversity 96: 1–242. https://doi.org/10.1007/s13225-019-00429-2
- Jiang, H.B., Phookamsak, R., Hyde, K.D., Mortimer, P.E., Xu, J.C., Kakumyan, P., Karunarathna, S.C. & Kumla, J. (2021) A taxonomic appraisal of bambusicolous fungi in Occultibambusaceae (Pleosporales, Dothideomycetes) with new collections from Yunnan Province, China. Life 11 (9): 932.
- Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/mswo54
- Li, X.W., Xiong, Z.P., Zhou, Y.P., Wang, J.H., Wei, J.F., Chen, X.X. & Xiong, Z. (2021) Study on Diversity of Culturable Rock-Inhabiting Actinomycetes in Shilin Rocky Desertification Area. Journal of Southwest University Natural Science Edition 43 (2): 70–78. https://doi.org/10.13718/j.cnki.xdzk.2021.02.010
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes evidence from an RNApolymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Lv, R., Yang, X., Qiao, M., Fang, L., Li, J. & Yu, Z. (2022) Exophiala yunnanensis and Exophiala yuxiensis (Chaetothyriales, Herpotrichiellaceae), two new species of soil-inhabiting Exophiala from Yunnan Province, China. Mycokeys 94: 109. https://doi.org/10.3897/mycokeys.94.96782
- Magaña-Dueñas, V., Stchigel, A.M. & Cano-Lira, J.F. (2021) New Coelomycetous Fungi from Freshwater in Spain. Journal of Fungi 7 (5). https://doi.org/10.3390/jof7050368
- Mapook, A., Hyde, K.D., McKenzie, E.H., Jones, E.G., Bhat, D.J., Jeewon, R., Stadler, M., Samarakoon, M.C., Malaithong, M., Tanunchai, B., Buscot, F., Wubet, T. & Purahong, W. (2020) Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Diversity 101: 1–175. https://doi.org/10.1007/s13225-020-00444-8
- Papizadeh, M., Soudi, M., Amini, L., Wijayawardene, N. & Hyde, K. (2017) Pyrenochaetopsis tabarestanensis (Cucurbitariaceae, Pleosporales), a new species isolated from rice farms in north Iran. Phytotaxa 297: 15–28. https://doi.org/10.11646/phytotaxa.297.1.2
- Phookamsak, R., Jiang, H., Suwannarach, N., Lumyong, S., Xu, J.C., Xu, S., Liao, C.F. & Chomnunti, P. (2022) Bambusicolous fungi in Pleosporales: Introducing four novel taxa and a new habitat record for Anastomitrabeculia didymospora. Journal of Fungi 8 (6): 630. https://doi.org/10.3390/jof8060630
- Qiao, M., Zhang, Z., Yang, L. & Yu, Z. (2019) Staphylotrichum sinense sp. nov., a new hyphomycete (Chaetomiaceae) from China. International Journal of Systematic and Evolutionary Microbiology 71 (3). https://doi.org/10.1099/ijsem.0.004747
- Rambaut, A. (2012) FigTree v1.4.2. [http://tree.bio.ed.ac.uk/]
- Rayner, R.W. (1970) A Mycological Colour Chart. Kew Commonwealth Mycological Institute. https://doi.org/10.1111/j.1477-9552.1970.tb02033.x
- Rehner, S.A. & Samuels, G.J. (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear largesubunit ribosomal DNA sequences. Mycological Research 98 (6): 625–634. https://doi.org/10.1016/S0953-7562(09)80409-7
- Samarakoon, B.C., Wanasinghe, D.N., Samarakoon, M.C., Bundhun, D., Jayawardena, R., Hyde, K.D. & Chomnunti, P. (2024) Exploring fungi: a taxonomic and phylogenetic study of leaf-inhabiting Ascomycota in Musa species from northern Thailand, with a global checklist. Mycosphere 15 (1): 1901–2174. https://doi.org/10.5943/mycosphere/15/1/17
- Špetík, M., Aberraf-Tebbal, A., Pokluda, R. & Eichmeier, A. (2021) Pyrenochaetopsis kuksensis (Pyrenochaetopsidaceae), a new species associated with an ornamental boxwood in the Czech Republic. Phytotaxa 498 (3): 177–185. https://doi.org/10.11646/phytotaxa.498.3.3
- Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford,England) 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446
- Sutton, B.C. (1980) The Coelomycetes. fungi imperfecti with pycnidia, acervuli and stromata. 696 pp.
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. & Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Acids Research 25: 4876–4882. https://doi.org/10.1093/nar/25.24.4876
- Valenzuela-Lopez, N., Cano-Lira, J.F., Guarro, J., Sutton, D.A., Wiederhold, N., Crous, P.W. & Stchigel, A.M. (2018) Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Studies in Mycology 90: 1–69. https://doi.org/10.1016/j.simyco.2017.11.003
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymaticallyamplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172 (8): 4238–4246.
- Wang, K.Y., Wu, Y.M., Chen, Y. & Jiang, Y.L. (2019) A new species of Pyrenochaetopsis and a key to the known species of the genus. Mycosystema 38 (2): 171–177.
- White, T.J., Bruns, T., Lee, S.J. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. Academic Press, USA, pp. 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1
- Woudenberg, J.H.C., Aveskamp, M.M., De Gruyter, J., Spiers, A.G. & Crous, P.W. (2009) Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia-Molecular Phylogeny and Evolution of Fungi 22 (1): 56–62. https://doi.org/10.3767/003158509x427808
- Yang, X.Q., Feng, M.Y. & Yu, Z.F. (2021) Exophiala pseudooligosperma sp. nov., a novel black yeast from soil in southern China. International Journal of Systematic Evoltionary Microbioogy 71 (11): 005116. https://doi.org/10.3897/mycokeys.94.96782
- Yang, X., Lv, R., Yu, Z., Li, J. & Qiao, M. (2022) Cordana yunnanensis sp. nov., Isolated from Desertified Rocky Soil in Southwest China. Current Microbiology 79 (6): 183. https://doi.org/10.1007/s00284-022-02871-z
- Zheng, H., Qiao, M., Lv, Y.F., Du, X., Zhang, K.Q. & Yu, Z.F. (2021) New species of soil-inhabiting and aquatic endophytic Trichoderma from southwest China. Journal of Fungi 7 (467): 10–3390. https://doi.org/10.3390/jof7060467