Abstract
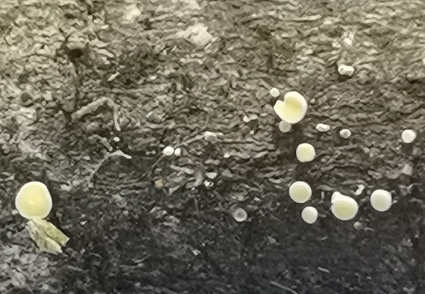
Proliferodiscus is a discomycete genus belonging to the family Lachnaceae (Helotiales). In the present study, a new Proliferodiscus species is identified and introduced based on morphological characteristics and multi-loci phylogenetic analyses of the combined ITS and LSU sequences. Proliferodiscus ailaoensis sp. nov. is characterized by yellow to creamy apothecia, a yellow to white receptacle surface with bushy hairs, filiform, aseptate, unbranched paraphyses with a rounded apex, eight-spored, J- asci with a tapered pedicel, and ascospores that are narrowly fusiform with acute ends, 4–6-septate, and smooth-walled. An updated key to the known species of Proliferodiscus is also provided.
References
- Bien, S. & Damm, U. (2020) Arboricolonus simplex gen. et sp. nov. and novelties in Cadophora, Minutiella and Proliferodiscus from Prunus wood in Germany. MycoKeys 63: 119–161. https://doi.org/10.3897/mycokeys.63.46836
- Cantrell, S.A. & Haines, J.H. (1997) New red species of Lachnum from the tropics. Mycological Research 101: 1081–1084. https://doi.org/10.1017/S0953756297003699
- Cantrell, S.A. & Hanlin, R.T. (1997) Phylogenetic relationships in the family Hyaloscyphaceae inferred from sequences of ITS regions, 5.8 S ribosomal DNA and morphological characters. Mycologia 89 (5): 745–755. https://doi.org/10.1080/00275514.1997.12026841
- Chaiwan, N., Gomdola, D., Wang, S., Monkai, J., Tibpromma, S., Doilom, M., Wanasinghe, D., Mortimer, P., Lumyong, S. & Hyde, K.D. (2021) An online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere 12: 1513–1526. https://doi.org/10.5943/mycosphere/12/1/19
- Chethana, K.W.T, Manawasinghe, I.S, Hurdeal, V.G, Bhunjun, C.S., Appadoo, M., Gentekaki, E., Raspé, O., Promputtha, I. & Hyde, K.D. (2021) What are fungal species and how to delineate them? Fungal Diversity 109: 1–25. https://doi.org/10.1007/s13225-021-00483-9
- Ekanayaka, A.H., Hyde, K.D., Gentekaki, E., McKenzie, E.H.C., Zhao, Q., Bulgakov, T.S. & Camporesi, E. (2019) Preliminary classification of Leotiomycetes. Mycosphere 10 (1): 310–489. https://doi.org/10.5943/mycosphere/10/1/7
- Gardes, M. & Bruns, T.D. (1993) ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. https://doi.org/10.1093/sysbio/syq010
- Guo, M.J., Zhuo, L., Wang, S.J., Sui, X.N., Zhou, H., Cai, S.R., Luo, J.T., Lei, R.H., Shen, X.Y., Piepenbring, M. & Hou, C.L. (2024) Hyperdiverse Rhytismatales on twigs of Rhododendron spp. Mycosphere 15 (1): 764–880. https://doi.org/10.5943/mycosphere/15/1/6
- Ekanayaka, A.H., Hyde, K.D., Gentekaki, E., McKenzie, E.H.C., Zhao, Q., Bulgakov, T.S. & Camporesi, E. (2019) Preliminary classification of Leotiomycetes. Mycosphere 10: 310–489. https://doi.org/10.5943/mycosphere/10/1/7
- Haines, J. & Dumont, K. (1983) Studies in the Hyaloscyphaceae II: Proliferodiscus, a new genus of Arachnopezizoideae. Mycologia 75 (3): 535–543. https://doi.org/10.1080/00275514.1983.12023717
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. https:// doi.org/10.14601/phytopathol_mediterr-14998u1.29
- Haelewaters, D., Park, D. & Johnston, P.R. (2021) Multilocus phylogenetic analysis reveals that Cyttariales is a synonym of Helotiales. Mycological Progress 20 (10): 1323–1330. https://doi.org/10.1007/s11557-021-01736-2
- Hamelin, R.C., Tanguay, P., Uzunovic, A.A. & Seifert, K.A. (2013) Molecular detection assays of forest pathogens. Canadian Forest Service. Natural Resources Canada. [http://getentry.ddbj.nig.ac.jp/getentry/na/KC464626]
- Hosoya, T., Sasagawa, R., Hosaka, K., Gi-Ho, S., Hirayama, Y., Yamaguchi, K., Toyama, K. & Kakishima, M. (2010) Molecular phylogenetic studies of Lachnum and its allies based on the Japanese material. Mycoscience 51 (3): 170–181. https://doi.org/10.1007/S10267-009-0023-1
- Hoang, D.T., Chernomor, O., Von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281
- Hyde, K.D., Noorabadi, M.T., Thiyagaraja, V., He, M.Q., Johnston, P.R., Wijesinghe, S.N., Armand, A., Biketova, A.Y., Chethana, K.W.T., Erdoğdu, M., Ge, Z.W., Groenewald, J.Z., Hongsanan, S., Kušan, I., Leontyev, D.V., Li, D.W., Lin, C.G., Liu, N.G., Maharachchikumbura, S.S.N., Matočec, N., May, T.W., McKenzie, E.H.C., Mešić, A., Perera, R.H., Phukhamsakda, C., Piątek, M., Samarakoon, M.C., Selcuk, F., Senanayake, I.C., Tanney, J.B., Tian, Q., Vizzini, A., Wanasinghe, D.N., Wannasawang, N., Wijayawardene, N.N., Zhao, R.L., Abdel-Wahab, M.A., Abdollahzadeh, J., Abeywickrama, P.D., Abhinav, Absalan, S., Acharya, K., Afshari, N., Afshan, N.S., Afzalinia, S., Ahmadpour, S.A., Akulov, O., Alizadeh, A., Alizadeh, M., Al-Sadi, A.M., Alves, A., Alves, V.C.S., Alves-Silva, G., Antonín, V., Aouali, S., Aptroot, A., Apurillo, C.C.S., Arias, R.M., Asgari, B., Asghari, R., Assis, D.M.A., Assyov, B., Atienza, V., Aumentado, H.D.R., Avasthi, S., Azevedo, E., Bakhshi, M., Bao, D.F., Baral, H.O., Barata, M., Barbosa, K.D., Barbosa, R.N., Barbosa, F.R., Baroncelli, R., Barreto, G.G., Baschien, C., Bennett, R.M., Bera, I., Bezerra, J.D.P., Bhunjun, C.S., Bianchinotti, M.V., Błaszkowski, J., Boekhout, T., Bonito, G.M., Boonmee, S., Boonyuen, N., Bortnikov, F.M., Bregant, C., Bundhun, D., Burgaud, G., Buyck, B., Caeiro, M.F., Cabarroi-Hernández, M., Cai, M.F., Cai, L., Calabon, M.S., Calaça, F.J.S., Callalli, M., Câmara, M.P.S., Cano-Lira, J., Cao, B., Carlavilla, J.R., Carvalho, A., Carvalho, T.G., Castañeda-Ruiz, R.F., Catania, M.D.V., Cazabonne, J., Cedeño-Sanchez, M., Chaharmiri-Dokhaharani, S., Chaiwan, N., Chakraborty, N., Cheewankoon, R., Chen, C., Chen, J., Chen, Q., Chen, Y.P., Chinaglia, S., Coelho-Nascimento, C.C., Coleine, C., Costa-Rezende, D.H., Cortés-Pérez, A., Crouch, J.A., Crous, P.W., Cruz, R.H.S.F., Czachura, P., Damm, U., Darmostuk, V., Daroodi, Z., Das, K., Das, K., Davoodian, N., Davydov, E.A., da Silva, G.A., da Silva, I.R., da Silva, R.M.F., da Silva, S.A.C., Dai, D.Q., Dai, Y.C., de Groot Michiel, D., De Kesel, A., De Lange, R., de Medeiros, E.V., de Souza, C.F.A., de Souza, F.A., dela Cruz, T.E.E., Decock, C., Delgado, G., Denchev, C.M., Denchev, T.T., Deng, Y.L., Dentinger, B.T.M., Devadatha, B., Dianese, J.C., Dima, B., Doilom, M., Dissanayake, A.J., Dissanayake, D.M.L.S., Dissanayake, L.S., Diniz, A.G., Dolatabadi, S., Dong, J.H., Dong, W., Dong, Z.Y., Drechsler-Santos, E.R., Druzhinina, I.S., Du, T.Y., Dubey, M.K., Dutta, A.K., Elliott, T.F., Elshahed, M.S., Egidi, E., Eisvand, P., Fan, L., Fan, X., Fan, X.L., Fedosova, A.G., Ferro, L.O., Fiuza, P.O., Flakus, A., W. Fonseca, E.O., Fryar, S.C., Gabaldón, T., Gajanayake, A.J., Gannibal, P.B., Gao, F., García-Sánchez, D., García-Sandoval, R., Garrido-Benavent, I., Garzoli, L., Gasca-Pineda, J., Gautam, A.K., Gené, J., Ghobad-Nejhad, M., Ghosh, A., Giachini, A.J., Gibertoni, T.B., Gentekaki, E., Gmoshinskiy, V.I., Góes-Neto, A., Gomdola, D., Gorjón, S.P., Goto, B.T., Granados-Montero, M.M., Griffith, G.W., Groenewald, M., Grossart, H-P., Gu, Z.R., Gueidan, C., Gunarathne, A., Gunaseelan, S., Guo, S.L., Gusmão, L.F.P., Gutierrez, A.C., Guzmán-Dávalos, L., Haelewaters, D., Haituk, H., Halling, R.E., He, S.C., Heredia, G., Hernández-Restrepo, M., Hosoya, T., Hoog, S.D., Horak, E., Hou, C.L., Houbraken, J., Htet, Z.H., Huang, S.K., Huang, W.J., Hurdeal, V.G., Hustad, V.P., Inácio, C.A., Janik, P., Jayalal, R.G.U., Jayasiri, S.C., Jayawardena, R.S., Jeewon, R., Jerônimo, G.H., Jin, J., Jones, E.B.G., Joshi, Y., Jurjević, Ž., Justo, A., Kakishima, M., Kaliyaperumal, M., Kang, G.P., Kang, J.C., Karimi, O., Karunarathna, S.C., Karpov, S.A., Kezo, K., Khalid, A.N., Khan, M.K., Khyaju, S., Kirchmair, M., Klawonn, I., Kraisitudomsook, N., Kukwa, M., Kularathnage, N.D., Kumar, S., Lachance, M.A., Lado, C., Latha, K.P.D., Lee, H.B., Leonardi, M., Lestari, A.S., Li, C., Li, H., Li, J., Li, Q., Li, Y., Li, Y.C., Li, Y.X., Liao, C.F., Lima, J.L.R., Lima, J.M.S., Lima, N.B., Lin, L., Linaldeddu, B.T., Linn, M.M., Liu, F., Liu, J.K., Liu, J.W., Liu, S., Liu, S.L., Liu, X.F., Liu, X.Y., Longcore, J.E., Luangharn, T., Luangsa-ard, J.J., Lu, L., Lu, Y.Z., Lumbsch, H.T., Luo, L., Luo, M., Luo, Z.L., Ma, J., Madagammana, A.D., Madhushan, A., Madrid, H., Magurno, F., Magyar, D., Mahadevakumar, S., Malosso, E., Malysh, J.M., Mamarabadi, M., Manawasinghe, I.S., Manfrino, R.G., Manimohan, P., Mao, N., Mapook, A., Marchese, P., Marasinghe, D.S., Mardones, M., Marin-Felix, Y., Masigol, H., Mehrabi, M., Mehrabi-Koushki, M., Meiras-Ottoni, A.de., Melo, R.F.R., Mendes-Alvarenga, R.L., Mendieta, S., Meng, Q.F., Menkis, A., Menolli, Jr.N., Mikšík, M., Miller, S.L., Moncada, B., Moncalvo, J.M., Monteiro, J.S., Monteiro, M., Mora-Montes, H.M., Moroz, E.L., Moura, J.C., Muhammad, U., Mukhopadhyay, S., Nagy, G.L., Najam, ul.Sehar.A., Najafiniya, M., Nanayakkara, C.M., Naseer, A., Nascimento, E.C.R,, Nascimento, S.S., Neuhauser, S., Neves, M.A., Niazi, A.R., Nie, Y., Nilsson, R.H., Nogueira, P.T.S., Novozhilov, Y.K., Noordeloos, M., Norphanphoun, C., Nuñez Otaño, N., O’Donnell, R.P., Oehl, F., Oliveira, J.A., Oliveira Junior, I., Oliveira, N.V.L., Oliveira, P.H.F., Orihara, T., Oset, M., Pang, K.L., Papp, V., Pathirana, L.S., Peintner, U., Pem, D., Pereira, O.L., Pérez-Moreno, J., Pérez-Ortega, S., Péter, G., Pires-Zottarelli, C.L.A., Phonemany, M., Phongeun, S., Pošta, A., Prazeres, J.F.S.A., Quan, Y., Quandt, C.A., Queiroz, M.B., Radek, R., Rahnama, K., Raj, K.N.A., Rajeshkumar, K.C, Rajwar, S., Ralaiveloarisoa, A.B., Rämä, T., Ramírez-Cruz, V., Rambold, G., Rathnayaka, A.R., Raza, M., Ren, G.C., Rinaldi, A.C., Rivas-Ferreiro, M., Robledo, G.L., Ronikier, A., Rossi, W., Rusevska, K., Ryberg, M., Safi, A., Salimi, F., Salvador-Montoya, C.A., Samant, B., Samaradiwakara, N.P., Sánchez-Castro, I., Sandoval-Denis, M., Santiago, A.L.C.M.A., Santos, A.C.D.S., Santos, L.A.dos., Sarma, V.V., Sarwar, S.. Savchenko, A., Savchenko, K., Saxena, R.K., Schoutteten, N., Selbmann, L., Ševčíková, H., Sharma, A., Shen, H.W., Shen, Y.M., Shu, Y.X., Silva, H.F., Silva-Filho, A.G.S., Silva, V.S.H., Simmons, D.R., Singh, R., Sir, E.B., Sohrabi, M., Souza, F.A., Souza-Motta, C.M., Sri-indrasutdhi, V., Sruthi, O.P., Stadler, M., Stemler, J., Stephenson, S.L., Stoyneva-Gaertner, M.P., Strassert, J.F.H., Stryjak-Bogacka, M., Su, H., Sun, Y.R., Svantesson, S., Sysouphanthong, P., Takamatsu, S., Tan, T.H., Tanaka, K., Tang, C., Tang, X., Taylor, J.E., Taylor, P.W.J., Tennakoon, D.S., Thakshila, S.A.D., Thambugala, K.M., Thamodini, G.K., Thilanga, D., Thines,M., Tiago, P.V., Tian, X.G., Tian, W.H., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Tomšovský, M., Torruella, G., Tsurykau, A., Udayanga, D., Ulukapı, M., Untereiner, W.A., Usman, M., Uzunov, B.A., Vadthanarat, S., Valenzuela, R., Van, den, Wyngaert, S., Van, Vooren, N., Velez, P., Verma, R.K., Vieira, L.C. Vieira, W.A.S., Vinzelj, J.M., Tang, A.M.C., Walker, A., Walker, A.K., Wang, Q.M., Wang, Y., Wang, X.Y., Wang, Z.Y., Wannathes, N., Wartchow, F., Weerakoon, G., Wei, D.P., Wei, X., White, J.F., Wijesundara, D.S.A., Wisitrassameewong, K., Worobiec, G., Wu, H.X., Wu, N., Xiong, Y.R., Xu, B., Xu, J.P., Xu, R., Xu, R.F., Xu, R.J., Yadav, S., Yakovchenko, L.S., Yang, H.D., Yang, X., Yang, Y.H., Yang, Y., Yang, Y.Y., Yoshioka, R., Youssef, Noha, H., Yu, F.M., Yu, Z.F., Yuan, L.L., Yuan, Q., Zabin, D.A., Zamora, J.C., Zapata, C.V., Zare, R., Zeng, M., Zeng, X.Y., Zhang, J.F., Zhang, J.Y., Zhang, S., Zhang, X.C., Zhao, C.L., Zhao, H., Zhao, Q., Zhao, H., Zhao, H.J., Zhou, H.M., Zhu, X.Y., Zmitrovich, I.V., Zucconi, L. & Zvyagina, E. (2024a) The 2024 Outline of Fungi and fungus-like taxa. Mycosphere 15 (1): 5146–6239. https://doi 10.5943/mycosphere/15/1/25
- Hyde, K.D., Saleh, A., Aumentado, H.D.R., Boekhout, T., Bera, I., Khyaju, S., Bhunjun, C.S., Chethana, K.W.T., Phukhamsakda, C., Doilom, M., Thiyagaraja, V., Mortimer, P.E., Maharachchikumbura, S.S.N., Hongsanan, S., Jayawardena, R.S., Dong, W., Jeewon, R., Al‑Otibi, F.,Wijesinghe, S.N. & Wanasinghe, D.N. (2024b) Fungal numbers: global needs for a realistic assessment. Fungal Diversity 128: 191–225. https://doi.org/10.1007/s13225-024-00545-8
- Jaklitsch, W.M., Baral, H.O., Lücking, R. & Lumbsch, H.T. (2016) Ascomycota. In: Frey, W. (Ed.) Syllabus of plant families—Adolf Engler’s syllabus der pfanzenfamilien. Borntraeger, Stuttgart, pp 1–322
- Johnston, P.R. & Baschien, C. (2020) Tricladiaceae fam. nov. (Helotiales, Leotiomycetes). Fungal Systematics and Evolution 6 (1): 233–242. https://doi.org/10.3114/fuse.2020.06.10
- Jayasiri, S.C., Hyde, K.D., Ariyawansa, H.A., Bhat, J., Buyck, B., Cai, L., Dai, Y.C., Abd-Elsalam, K.A., Ertz, D., Hidayat, I., Jeewon, R., Jones, E.B.G., Bahkali, A.H., Karunarathna, S.C., Liu, J.K., Luangsa-ard, J.J., Lumbsch, H.T., Maharachchikumbura, S.S.N., McKenzie, E.H.C., Moncalvo, J.M., Ghobad-Nejhad, M., Nilsson, H., Pang, K.L., Pereira, O.L., Phillips, A.J.L., Raspé, O., Rollins, A.W., Romero, A.I., Etayo, J., Selçuk, F., Stephenson, S.L., Suetrong, S., Taylor, J.E., Tsui, C.K.M., Vizzini, A., Abdel-Wahab, M.A., Wen, T.C., Boonmee, S., Dai, D.Q., Daranagama, D.A., Dissanayake, A.J., Ekanayaka, A.H., Fryar, S.C., Hongsanan, S., Jayawardena, R.S., Li, W.J., Perera, R.H., Phookamsak, R., de Silva, N.I., Thambugala, K.M., Tian, Q., Wijayawardene, N.N., Zhao, R.L., Zhao, Q., Kang, J.C. & Promputtha, I. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74 (1): 3–18. https://doi.org/10.1007/s13225-015-0351-8
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K., Von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010
- Li, C.J.Y., Chethana, K.W.T., Lu, Z.Y. & Zhao, Q. (2022) Two novel species of Lachnaceae (Helotiales, Leotiomycetes) from southwestern China. Current Research in Environmental & Applied Mycology (Journal of Fungal Biology) 12: 333–345. https://doi.org/10.5943/cream/12/1/20
- Li, C.J.Y., Chethana, K.W.T., Eungwanichayapant, P.D., Luo, L., Yang, Z.L., Dong, W.J., Guo, Y.Y., Liu, C.H., Alotibi, F., Hyde, K.D. & Zhao, Q. (2024) Longistipes gen. nov. and four novel species of Hyphodiscaceae along with six new collections of Leotiomycetes in Yunnan Province, China. Mycosphere 15 (1): 4744–4787. https://doi.org/10.5943/mycosphere/15/1/20
- Luo, L., Wei, D.P., Zhao, Q. & Chethana, K.W.T. (2024) Unveiling the Diversity: A novel species of Dicephalospora (Helotiaceae, Helotiales) discovered in China. Phytotaxa 652: 59–68. https://doi.org/10.11646/phytotaxa.652.1.5
- Maharachchikumbura, S.S.N., Chen, Y., Ariyawansa, H.A., Hyde, K.D., Haelewaters, D., Perera, R.H., Samarakoon, M.C., Wanasinghe, D.N., Bustamante, D.E., Liu, J., Lawrence, D.P., Cheewangkoon, R. & Stadler, M. (2021) Integrative approaches for species delimitation in Ascomycota. Fungal Diversity 109: 155–179. https://doi.org/10.1007/s13225-021-00486-6
- Markéta, Š., Henrik, N.R. & Kolařík, M. (2018) Relationships within Capitotricha bicolor (Lachnaceae, Ascomycota) as inferred from ITS rDNA sequences, including some notes on the Brunnipila and Erioscyphella clades. Mycological Progress 17: 89–101. https://doi.org/10.1007/s11557-017-1346-5
- McKenzie, E.H.C., Johnston, P.R. & Buchanan, P.K. (2006) Checklist of fungi on teatree (Kunzea and Leptospermum species) in New Zealand. New Zealand Journal of Botany 44 (3): 293–335. https://doi.org/10.1080/0028825X.2006.9513025
- Nguyen, L.T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. https://doi.org/10.1093/molbev/msu300
- Popov, Е.С. (2013) New records of some rare species in the family Hyaloscyphaceae (Ascomycota, Helotiales) from central Russia. Novosti Sistematiki Nizshikh Ras 47: 135–142. https://doi.org/10.31111/nsnr/2013.47.135
- Quandt, C.A. & Haelewaters, D. (2021) Phylogenetic advances in Leotiomycetes, an understudied clade of taxonomically and ecologically diverse fungi. Encyclopedia of Mycology 1: 284–294. https://doi.org/10.1016/B978-0-12-819990-9.00052-4
- Quijada, L., Matočec, N., Kušan, I., Tanney, J.B., Johnston, P.R., Mešićet, A. & Pfister, D.H. (2022) Apothecial ancestry, evolution, and re-evolution in Thelebolales (Leotiomycetes, Fungi). Biology 11 (4): 583. https://doi.org/10.3390/biology11040583
- Raitviir, A.G. (2004) Revised synopsis of the Hyaloscyphaceae. Scripta Mycologica 20: 1–133.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Spooner, B.M. (1987) Helotiales. of Australasia: Geoglossaceae, Orbiliaceae, Sclerotiniaceae, Hyaloscyphaceae. Bibliotheca Mycologica 116: 470–474.
- Su, H.L., Chethana, K.W.T., Zeng, M. & Zhao, Q. (2023) Two new species of Erioscyphella (Lachnaceae) from southwestern. Current Research in Environmental & Applied Mycology 13 (1): 16–33. https://doi.org/10.5943/cream/13/1/2
- Swindell, S.R. & Plasterer, T. (1997) Sequence data analysis guidebook. Vol. 70. New York, NY, USA: Humana Press. https://doi.org/10.1385/0896033589
- Thiyagaraja, V., He, S.C., Luo, L., Meng, Q.F., Yang, H.D. & Yang, Y.Y. (2024) Research profiles–I: the research of Vinodhini Thiyagaraja and her team at Kunming Institute of Botany, CAS, PR China. Current Research in Environmental & Applied Mycology 14 (1): pp.49-62. https://doi.org/10.5943/cream/14/1/2
- Tochihara, Y. & Hosoya, T. (2019) Three new species of Incrucipulum (Lachnaceae, Helotiales, Ascomycota) from Japan. Phytotaxa 403 (1): 25–38. https://doi.org/10.11646/phytotaxa.403.1.2
- Tochihara, Y & Hosoya, T. (2022) Examination of the generic concept and species boundaries of the genus Erioscyphella (Lachnaceae, Helotiales, Ascomycota) with the proposal of new species and new combinations based on the Japanese materials. MycoKeys 87: 1–52. https://doi.org/10.3897/mycokeys.87.73082
- Tom, W., Taylor, A.F.S., Tobias, G.F., Julia, P., Björn, L. & Matthew, E.S. (2024) SH0869244.10FU. UNITE Community. https://dx.doi.org/10.15156/BIO/SH0869244.10FU
- Haelewaters, D., Dirks, A.C., Kappler, L.A., Mitchell, J.K., Quijada, L., Vandegrift, R. & Pfister, D.H. (2018) A preliminary checklist of fungi at the Boston Harbor Islands. Northeastern Naturalist 25 (sp9): 45–76. https://doi.org/10.1656/045.025.s904
- Hamelin, R.C., Tanguay, P., Uzunovic, A.A. & Seifert, K.A. (2013) Molecular detection assays of forest pathogens. Canadian Forest Service. Natural Resources Canada. http://getentry. ddbj. nig. ac. jp/getentry/na/KC464626.
- Han, J.G., Hosoya, T., Sun, G.H. & Shin, H.D. (2014) Phylogenetic reassessment of Hyaloscyphaceae sensu lato (Helotiales, Leotiomycetes) based on multigene analyses. Fungal Biology 118 (2): 150–167. https://doi.org/10.1016/j.funbio.2013.11.004
- Hosoya, T., Sasagawa, R., Hosaka, K., Gi-Ho, S., Hirayama, Y., Yamaguchi, K., Toyama, K. & Kakishima, M. (2010) Molecular phylogenetic studies of Lachnum and its allies based on the Japanese material. Mycoscience 51 (3): 170–181. https://doi.org/10.1007/S10267-009-0023-1
- Hosoya, T., Zhao, Y.J., Han, J.G., Saito, Y. & Kakishima, M. (2012) Enumeration of remarkable Japanese discomycetes (6): notes on two inoperculate discomycetes new to Japan and one operculate discomycete. Bulletin of the National Museum of Nature and Science, Series B, 38: 139–146.
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- Urmas, K., Kessy, A., Henrik, N.R., Karl-Henrik, L., Tom, W.M., Taylor, A.F.S., Tobias, G.F. & Kadri, P. (2024) TH108813. UNITE Community. https://10.15156/BIO/TH108813
- Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Groenewald, J.Z., Cardinali, G., Houbraken, J., Boekhout, T., Crous, W., Robert, V. & Verkley, G.J.M. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92 (1): 135–154. https://doi.org/10.1016/j.simyco.2018.05.001
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sánchez-García, M., Goto, B.T., Saxena, R.K., Erdoğdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., Błaszkowski, J., Boonyuen, N., da Silva, G.A., De Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Mešić, A., Monteiro, J.S., Oehl, F., Pawłowska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro, I., Tang, L.Z., Stadler, M., Yurkov, A. & Thines, M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13 (1): 53–453. https://doi.org/10.5943/mycosphere/13/1/2
- Wu, W. & Diao, Y. (2023) The chalara-like anamorphs of Leotiomycetes. Fungal Diversity 119 (1): 213–490. https://doi.org/10.1007/s13225-023-00515-6
- Ye, M., Cao, S.Q., Jiang, S.T., Pan, L.J., Luo, S.Z. & Li, X.J. (2006) Species diversity of Lachnum (Helotiales, Hyaloscyphaceae) from temperate China. Journal of Zhejiang University Science B 7 (1): 20–27. https://doi.org/10.1631/jzus.2006.B0020
- Zeng, M., Gentekaki, E., Zeng, X.Y., Tian, Q., Zhao, Q. & Hyde, K.D. (2022) Evolutionary relationships and allied species of Pyronemataceae, with segregation of the novel family Pyropyxidaceae. Mycosphere 13 (2): 207–280. https://doi.org/10.5943/mycosphere/si/1f/7
- Zeng, X.Y., Tan, T.J., Tian, F.H., Wang, Y. & Wen, T.C. (2023) OFPT: a one-stop software for fungal phylogeny. Mycosphere 14 (1): 1730–1741. https://doi.org/10.5943/mycosphere/14/1/20
- Zhao, Y.J., Hosoya, T., Baral, H.O., Hosaka, K. & Kakishima, M. (2013) Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon 122 (1): 25–41. https://doi.org/10.5248/122.25
- Zhang, Z.Y., Pan, H., Tao, G., Li, X., Han, Y.F., Feng, Y., Tong, S.Q. & Ding, C.Y. (2024) Culturable mycobiota from Guizhou Wildlife Park in China. Mycosphere 15 (1): 654–763. https://doi.org/10.1007/s11557-013-0945-z
- Zheng, H.D. & Zhuang, W.Y. (2014) Hymenoscyphus albidoides sp. nov. and H. pseudoalbidus from China. Mycological Progress 13: 625–638. https://doi.org/10.1007/s11557-013-0945-z
- Zhuang, W.Y. (2000) Hyaloscyphaceous discomycetes from Ningxia Province, China. Mycologia 92 (3): 593–597. https://doi.org/10.1080/00275514.2000.12061198
- Zhuang, W.Y. & Hyde, K.D. (2001) New species of Lachnum and Perrotia from Hong Kong, China. Mycologia 93 (3): 606–611. https://doi.org/10.1080/00275514.2001.12063190
- Zhuang, W.Y., Yu, Z.H., Zhang, Y.H. & Ming, Y. (2002) Some new species and new records of discomycetes in China. IX. Mycotaxon 81: 27–34.