Abstract
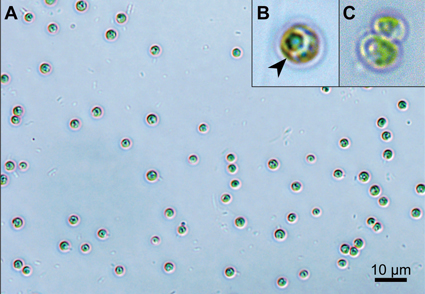
Green algae belong to the major photosynthetic organisms in aquatic environments. Their taxonomy has undergone dramatic revisions in recent decades, resulting in numerous new taxa. However, many small-celled spherical morphotypes have so far remained uncertain with regard to their classification. With the help of molecular phylogenetic methods, these taxa can be more clearly defined. In the present study, we isolated a freshwater green alga (SMU-GA001) from the Yun River, Republic of Korea. The species has spherical, solitary cells with a cup-shaped chloroplast without pyrenoids. The cell size ranged from 1.5 to 3.5 µm. Based on these morphological characteristics, we identified the species to be within the genus Mychonastes. The identity was confirmed by the phylogenetic analysis of 18S rRNA; however, the species differs from other Mychonastes species. The species identified here showed unique molecular features regarding the rRNA ITS2 sequence and its secondary structure, along with compensatory base changes (CBCs), in comparison to other Mychonastes species. These results revealed that our algae is a novel species within the genus Mychonastes, and thus we proposed it as Mychonastes koreanus sp. nov., named after Korea, the country where it was first discovered.
References
- Ankenbrand, M.J., Keller, A., Wolf, M., Schultz, J. & Förster, F. (2015) ITS2 database V: Twice as much. Molecular Biology and Evolution 32: 3030–3032. https://doi.org/10.1093/molbev/msv174
- Byun, Y. & Han, K. (2009) PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics 25: 1435–1437. https://doi.org/10.1093/bioinformatics/btp252
- Caisová, L., Marin, B. & Melkonian, M. (2013) A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 164: 482–496. https://doi.org/10.1016/j.protis.2013.04.005
- Coleman, A.W. (2000) The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151: 1–9. https://doi.org/10.1078/1434-4610-00002
- Coleman, A.W. (2007) Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Research 35: 3322–3329. https://doi.org/10.1093/nar/gkm233
- Darienko, T., Gustavs, L., Eggert, A., Wolf, W. & Pröschold, T. (2015) Evaluating the species boundaries of green microalgae (Coccomyxa, Trebouxiophyceae, Chlorophyta) using integrative taxonomy and DNA barcoding with further implications for the species identification in environmental samples. PLOS One 10 (6): e0127838. https://doi.org/10.1371/journal.pone.0127838
- Fawley, M.W., Fawley, K.P., Buchheim, J.A. & Buchheim, M.A. (2003) 45 Phylogeny and systematics of Pseudodictyosphaerium, Mychonastes and related coccoid green algae (Chlorophyceae). Journal of Phycology 39: 16. https://doi.org/10.1111/j.0022-3646.2003.03906001_45.x
- Fitch, D.H., Bugaj-Gaweda, B. & Emmons, S.W. (1995) 18S ribosomal RNA gene phylogeny for some Rhabditidae related to Caenorhabditis. Molecular Biology and Evolution 12: 346–358. https://doi.org/10.1093/oxfordjournals.molbev.a040207
- Floyd, G.L., Watanabe, S. & Deason, T.R. (1993) Comparative ultrastructure of the zoospores of eight species of Characium (Chlorophyta). Archiv für Protistenkunde 143: 63–73. https://doi.org/10.1016/S0003-9365(11)80275-7
- Fucíková, C. & Lewis, L.E. (2012) Intersection of Chlorella, Muriella and Bracteacoccus: Resurrecting the genus Chromochloris Kol et Chodat (Chlorophyceae, Chlorophyta). Fottea 12 (1): 83–93. https://doi.org/10.5507/fot.2012.007
- Guiry, M.D. & Guiry, G.M. (2021) AlgaeBase. World-wide electronic publication. National University of Ireland, Galway. Available from: http://www.algaebase.org/ (accessed 20 May 2024)
- Guiry, M.D. (2024) How many species of algae are there? A reprise. Four kingdoms, 14 phyla, 63 classes and still growing. Journal of Phycology 60 (2): 214–228. https://doi.org/10.1111/jpy.13431
- Kalina, T. & Punčochářová, M. (1987) Taxonomy of the subfamily Scotiellocystoideae Fott 1976 (Chlorellaceae, Chlorophyceae). Algological Studies 45: 473–521.
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010
- Krienitz, L., Takeda, H. & Hepperle, D. (1999) Ultrastructure, cell wall composition, and phylogenetic position of Pseudodictyosphaerium jurisii (Chlorococcales, Chlorophyta) including a comparison with other picoplanktonic green algae. Phycologia 38: 100–107. https://doi.org/10.2216/i0031-8884-38-2-100.1
- Krienitz, L., Wolf, M., Hegewald, E. & Hepperle, D. (2003) Systematics of coccoid green algae: morphology vs. 18S rRNA gene phylogeny. Journal of Phycology 38: 19–20. https://doi.org/10.1046/j.1529-8817.38.s1.57.x
- Krienitz, L. & Hepperle, D. (2003) The systematics of coccoid green algae: 18S rRNA gene sequence data versus morphology. Biologia 58: 437–446.
- Krienitz, L., Bock, C., Dadheech, P.K. & Pröschold, T. (2011) Taxonomic reassessment of the genus Mychonastes (Chlorophyceae, Chlorophyta) including the description of eight new species. Phycologia 50: 89–106. https://doi.org/10.2216/10-15.1
- Krivina, E.S. & Temraleeva, A.D. (2020) Identification problems and cryptic diversity of Chlorella-clade microalgae (Chlorophyta). Microbiology 89: 720–732. https://doi.org/10.1134/S0026261720060107
- Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
- Leliaert, F., Smith, D.R., Moreau, H., Herron, M.D., Verbruggen, H., Delwiche, C.F. & De Clerck, O. (2012) Phylogeny and molecular evolution of the green algae. Critical Reviews in Plant Sciences 31: 1–46. https://doi.org/10.1080/07352689.2011.615705
- Margulis, L., Hinkle, G., McKhann, H. & Moynihan, B. (1988) Mychonastes desiccatus Brown sp. nova (Chlorococcales, Chlorophyta) an intertidal alga forming achlorophyllous desiccation-resistant cysts. Algological Studies 78: 425–446.
- Martynenko, N., Gusev, E., Kapustin, D. & Kulikovskiy, M. (2022) A new cryptic species of the genus Mychonastes (Chlorophyceae, Sphaeropleales). Plants 11: 3363. https://doi.org/10.3390/plants11233363
- Müller, T., Philippi, N., Dandekar, T., Schultz, J. & Wolf, M. (2007) Distinguishing species. RNA 13: 1469–1472. https://doi.org/10.1261/rna.617107
- Ogedengbe, J.D., Hanner, R.H. & Barta, J.R. (2011) DNA barcoding identifies Eimeria species and contributes to the phylogenetics of coccidian parasites (Eimeriorina, Apicomplexa, Alveolata). International Journal for Parasitology 41: 843–850. https://doi.org/10.1016/j.ijpara.2011.03.007
- O’Kelly, C.J., Kurihara, A., Shipley, T.C. & Sherwood, A.R. (2010) Molecular assessment of Ulva spp. (Ulvophyceae, Chlorophyta) in the Hawaiian Islands. Journal of Phycology 46: 728–735. https://doi.org/10.1111/j.1529-8817.2010.00860.x
- Pascher, A. (1918) Von einer allen Algenreihen gemeinsamen Entwicklungsregel. Berichte der Deutschen Botanischen Gesellschaft 36: 390–409.
- Patova, E., Novakovskaya, I., Martynenko, N., Gusev, E. & Kulikovskiy, M. (2021) Mychonastes frigidus sp. nov. (Sphaeropleales/Chlorophyceae), a new species described from a mountain stream in the Subpolar Urals (Russia). Fottea 21: 8–15. https://doi.org/10.5507/fot.2020.012
- Richards, E., Reichardt, M. & Rogers, S. (1994) Preparation of genomic DNA from plant tissue. Current Protocols in Molecular Biology 27 1: 2–3. https://doi.org/10.1002/0471142727.mb0203s27
- Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
- Sakayama, H., Hara, Y., Arai, S., Sato, H. & Nozaki, H. (2004) Phylogenetic analyses of Nitella subgenus Tieffallenia (Charales, Charophyceae) using nuclear ribosomal DNA internal transcribed spacer sequences. Phycologia 43: 672–681. https://doi.org/10.2216/i0031-8884-43-6-672.1
- Schultz, J., Maisel, S., Gerlach, D., Müller, T. & Wolf, M. (2005) A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 11: 361–364. https://doi.org/10.1261/rna.7204505
- Seibel, P.N., Müller, T., Dandekar, T., Schultz, J. & Wolf, M. (2006) 4SALE-a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics 7: 1–7. https://doi.org/10.1186/1471-2105-7-498
- Simpson, P.D. & Van Valkenburg, S.D. (1978) The ultrastructure of Mychonastes ruminatus gen. et sp. nov., a new member of the Chlorophyceae isolated from brackish water. British Phycological Journal 13: 117–130. https://doi.org/10.1080/00071617800650131
- Tang, C.Q., Leasi, F., Obertegger, U., Kieneke, A., Barraclough, T.G. & Fontaneto, D. (2012) The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proceedings of the National Academy of Sciences 109: 16208–16212. https://doi.org/10.1073/pnas.1209160109
- Thompson, J.D., Higgins, D.G. & Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. https://doi.org/10.1093/nar/22.22.4673
- Watanabe, S. & Floyd, G.L. (1992) Comparative ultrastructure of zoospores with parallel basal bodies from the green algae Dictyochloris fragrans and Bracteacoccus sp. American Journal of Botany 79: 551–555. https://doi.org/10.1002/j.1537-2197.1992.tb14592.x