Abstract
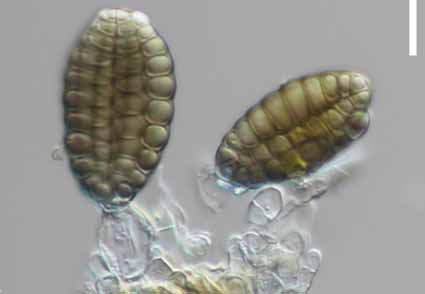
The diversity of lignicolous freshwater fungi in southwestern China is currently being studied. In this paper, two fungal isolates were collected from freshwater habitats in Yunnan Province, China. Based on morphological comparisons and the results of phylogenetic analysis of combined ITS, LSU, rpb2, SSU, and tef1-α sequence data, we introduce two new species, Aquadictyospora nujiangensis and Dendryphion verrucosum. Aquadictyospora nujiangensis is characterized by micronematous conidiophores and muriform, cheiroid conidia composed of compactly adpressed, curved rows of cells. Dendryphion verrucosum is characterized by erect, apically branched conidiophores, polytretic conidiogenous cells, and solitary or catenate to branched, monilioid conidia. This study contributes to the lignicolous freshwater fungal diversity in southwestern China.
References
- Bao, D.F., McKenzie, E.H., Bhat, D.J., Hyde, K.D., Luo, Z.L., Shen, H.W. & Su, H.Y. (2020) Acrogenospora (Acrogenosporaceae, Minutisphaerales) appears to be a very diverse genus. Frontiers in Microbiology 11: 1606. https://doi.org/10.3389/fmicb.2020.01606
- Bao, D.F., Hyde, K.D., McKenzie, E.H.C., Jeewon, R., Su, H.Y., Nalumpang, S. & Luo, Z.L. (2021) Biodiversity of lignicolous freshwater hyphomycetes from China and Thailand and description of sixteen species. Journal of Fungi 7: 669. https://doi.org/10.3390/jof7080669
- Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15): 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
- Calabon, M.S., Hyde, K.D., Jones, E.B.G., Luo, Z.L., Dong, W., Hurdeal, V.G., Gentekaki, E., Rossi, W., Leonardi, M., Thiyagaraja, V., Lestari, A.S., Shen, H.W., Bao, D.F., Boonyuen, N. & Zeng, M. (2022) Freshwater fungal numbers. Fungal Diversity 114 (1): 3–235. https://doi.org/10.1007/s13225-022-00503-2
- Calabon, M.S., Hyde, K.D., Jones, E.B.G., Bao, D.F., Bhunjun, C.S., Phukhamsakda, C., Shen, H.W., Gentekaki, E., Al Sharie, A.H., Barros, J., Chandrasiri, S.K.U., Hu, D.M., Hurdeal, V., Rossi, W., Valle, L.G., Zhang, H., Figueroa, M., Raja, H.A., Seena, S., Song, H.Y., Dong, W., El-Elimat, T., Leonardi, W., Li, Y., Li, Y.J., Luo, Z.L., Ritter, C.D., Strongman, D.B., Wei, M.J. & Balasuriya, A. (2023) Freshwater fungal biology. Mycosphere 14 (1): 195–413. https://doi.org/10.5943/mycosphere/14/1/4
- Crous, P.W., Shivas, R.G., Quaedvlieg, W., Bank, M., Zhang, Y., Summerell, B.A., Guarro, J., Wingfield, M.J., Wood, A.R., Alfenas, A.C., Braun, U., Cano-Lira, J.F., García, D., Marin-Felix, Y., Alvarado, P., Andrade, J.P., Armengol, J., Assefa, A., Breeÿen, A., Camele, I., Cheewangkoon, R., De Souza, J.T., Duong, T.A., Esteve-Raventós, F., Fournier, J., Frisullo, S., García-Jiménez, J., Gardiennet, A., Gené, J., Hernández-Restrepo, M., Hirooka, Y., Hospenthal, D.R., King, A., Lechat, C., Lombard, L., Mang, S.M., Marbach, P.A.S., Marincowitz, S., Marin-Felix, Y., Montaño-Mata, N.J., Moreno, G. Perez, C.A., Pérez Sierra, A.M., Robertson, J.L., Roux, J., Rubio, E., Schumacher, R.K., Stchigel, A.M., Sutton, D.A., Tan, Y.P., Thompson, E.H., Linde, E., Walker, A.K., Walker, D.M., Wickes, B.L., Wong, P.T.W. & Groenewald, J.Z. (2014) Fungal Planet description sheets: 214–280. Persoonia 32 (1): 184–306. https://doi.org/10.3767/003158514X682395
- Crous, P.W., Carris, L.M., Giraldo, A., Groenewald, J.Z., Hawksworth, D.L., Hernández-Restrepo, M., Jaklitsch, W.M., Lebrun, M., Schumacher, R.K., Stielow, J.B., van der Linde, E.J., Vilcāne, J., Voglmayr, H. & Wood, A.R. (2015) The Genera of Fungi-fixing the application of the type species of generic names–G2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6 (1): 163–198. https://doi.org/10.5598/imafungus.2015.06.01.11
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature methods 9 (8): 772–772. https://doi.org/10.1038/nmeth.2109
- Dong, W., Wang, B., Hyde, K.D., McKenzie, E.H., Raja, H.A., Tanaka, K., Abdel-Wahab, M.A., Abdel-Aziz, F.A., Doilom, M. & Phookamsak, R. (2020) Freshwater Dothideomycetes. Fungal Diversity 105: 319–575. https://doi.org/10.1007/s13225-020-00463-5
- Guindon, S. & Gascuel, O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology 52 (5): 696–704. https://doi.org/10.1080/10635150390235520
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. https://doi.org/10.12691/ajmr-6-4-3
- Hyde, K.D., Fryar, S., Tian, Q., Bahkali, A.H. & Xu, J.C. (2016) Lignicolous freshwater fungi along a northsouth latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecology 19: 190–200. https://doi.org/10.1016/j.funeco.2015.07.002
- Index Fungorum (2024) Index Fungorum. Available from: http://www.indexfungorum.org/ (accessed 4 December 2024)
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7 (11): 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010
- Li, J.F., Jeewon, R., Mortimer, P.E., Doilom, M., Phookamsak, R. & Promputtha, I. (2020a) Multigene phylogeny and taxonomy of Dendryphion hydei and Torula hydei spp. nov. from herbaceous litter in northern Thailand. PLoS One 15 (2): e0228067. https://doi.org/10.1371/journal.pone.0228067
- Li, L.L., Shen, H.W., Bao, D.F., Wanasinghe, D.N., Lu, Y.Z., Feng, Y. & Luo, Z.L. (2022) The plethora of Tubeufiaceae in lakes of the northwestern Yunnan plateau, China. Frontiers in Microbiology 13: 1056669. https://doi.org/10.3389/fmicb.2022.1056669
- Li, W.L., Luo, Z.L., Liu, J.K., Bhat, D.J., Bao, D.F., Su, H.Y. & Hyde, K.D. (2017) Lignicolous freshwater fungi from China I: Aquadictyospora lignicola gen. et sp. nov. and new record of Pseudodictyosporium wauense from northwestern Yunnan Province. Mycosphere 8: 1587–1597. https://doi.org/10.5943/mycosphere/8/10/1
- Li, W.L., Bao, D.F., Bhat, D.J. & Su, H.Y. (2020b) Tetraploa aquatica (Tetraplosphaeriaceae), a new freshwater fungal species from Yunnan Province, China. Phytotaxa 459: 181–189. https://doi.org/10.11646/phytotaxa.459.2.8
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Luan, S., Shen, H.W., Bao, D.F., Luo, Z.L. & Li, Y.X. (2023) Morphology and multi-gene phylogeny reveal a novel Torula (Pleosporales, Torulaceae) species from the plateau lakes in Yunnan, China. Biodiversity Data Journal 11: e109477. https://doi.org/10.3897/BDJ.11.e109477
- Luo, Z.L., Hyde, K.D., Liu, J.K., Bhat, D.J., Bao, D.F., Li, W.L., Su, X.J., Yang, X.Y. & Su, H.Y. (2018) Lignicolous freshwater fungi from China II: novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9: 444–461. https://doi.org/10.5943/mycosphere/9/3/2
- Luo, Z.L., Hyde, K.D., Liu, J.K., Maharachchikumbura, S.S.N., Jeewon, R., Bao, D.F., Bhat, D.J., Lin, C.G., Li, W.L., Yang, J. & Su, H.Y. (2019) Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. https://doi.org/10.1007/s13225-019-00438-1
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010. New Orleans, pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129
- Phukhamsakda, C., McKenzie, E.H.C., Phillips, A.J., Jones, E.B.G., Bhunjun, C.S., Wanasinghe, D.N., Thongbai, B., Camporesi, E., Ertz, D., Jayawardena, R.S., Perera, R.H., Ekanayake, A., Tibpromma, S., Doilom, M., Xu, J.C. & Hyde, K.D. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102: 1–203. https://doi.org/10.1007/s13225-020-00448-4
- Ranala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10.1007/BF02338839
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
- Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E. & Wanasinghe, D.N. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
- Shen, H.W., Bao, D.F., Wanasinghe, D.N., Boonmee, S., Liu, J.K. & Luo, Z.L. (2022) Novel Species and Records of Dictyosporiaceae from Freshwater Habitats in China and Thailand. Journal of Fungi 8: 1200. https://doi.org/10.3390/jof8111200
- Shen, H.W., Bao, D.F., Boonmee, S., Su, X.J., Tian, X.G., Hyde, K.D. & Luo, Z.L. (2023) Lignicolous Freshwater Fungi from Plateau Lakes in China (I): Morphological and Phylogenetic Analyses Reveal Eight Species of Lentitheciaceae, Including New Genus, New Species and New Records. Journal of Fungi 9 (10): 962. https://doi.org/10.3390/jof9100962
- Shen, H.W., Bao, D.F., Boonmee, S., Lu, Y.Z., Su, X.J., Li, Y.X. & Luo, Z.L. (2024) Diversity of Distoseptispora (Distoseptisporaceae) taxa on submerged decaying wood from the Red River in Yunnan, China. MycoKeys 102: 1–28. https://doi.org/10.3897/mycokeys.102.116096
- Shu, Y.X., Doilom, M., Boonmee, S., X, B. & Dong, W. (2024) Three novel cheiroid hyphomycetes in Dictyocheirospora and Dictyosporium (Dictyosporiaceae) from freshwater habitats in Guangdong and Guizhou Provinces, China. Journal of Fungi 10: 259. https://doi.org/10.3390/jof10040259
- Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446
- Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biology 75: 758–771. https://doi.org/10.1080/10635150802429642
- Su, H.Y., Hyde, K.D., Maharachchikumbura, S.S.N., Ariyawansa, H.A., Luo, Z.L., Promputtha, I., Tian, T., Lin, C.G., Shang, Q.J., Zhao, Y.C., Chai, H.M., Liu, X.Y., Bahkali, A.H., Bhat, D.J., McKenzie, E.H.C. & Zhou, D.Q. (2016) The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Diversity 80 (1): 375–409. https://doi.org/10.1007/s13225-016-0362-0
- Su, X.J., Luo, Z.L., Jeewon, R., Bhat, D.J., Bao, D.F., Li, W.L., Hao, Y.E. & Su, H.Y. & Hyde, K.D. (2018) Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan, China. Mycological Progress 17 (5): 531–545. https://doi.org/10.1007/s11557-018-1388-3
- Sung, G.H., Sung, J.M., Hywel-Jones, N.L. & Spatafora, J.W. (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Molecular Phylogenetics and Evolution 44 (3): 1204–1223. https://doi.org/10.1016/j.ympev.2007.03.011
- Vaidya, G., Lohman, D.J. & Meier, R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27 (2): 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- Wallroth, C.F.W. (1833) Flora Cryptogamica Germaniae. Vol. 2. 923 pp.
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. Academic Press: Cambridge, MA, USA, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Yang, J., Liu, L.L., Jones, E.B.G., Hyde, K.D., Liu, Z.Y., Bao, D.F., Liu, N.J., Li, W.L., Shen, H.W., Yu, X.D. & Liu, J.K. (2023) Freshwater fungi from karst landscapes in China and Thailand. Fungal Diversity 119: 1–212. https://doi.org/10.1007/s13225-023-00514-7