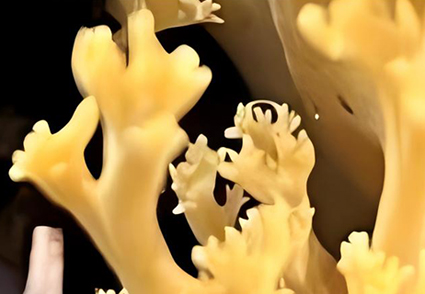
Abstract
During a survey of coral mushrooms in Yunnan Province, China, yellow, cylindrical, and coral-shaped basidiomata were collected and preliminarily identified as a Phaeoclavulina species. Phylogenetic analyses of combined ITS1–5.8S–ITS2 and LSU sequences support our collections positioning within Phaeoclavulina sister lineage to P. carovinacea strains with 58% BS and 0.95 PP statistical support. Our Phaeoclavulina collection is characterized by verrucose, truncate (volcanic) spines in spores, and clamp connections that differ from closely related species. Phylogenetic analysis results and morphological comparisons support our collection as a distinct new species of Phaeoclavulina (P. yunnanensis). The new species is described and illustrated, and the results of the phylogenetic analysis are provided.
References
- Beug, M. (2004) Trial field key to species of Ramaria in Pacific Northwest. (Cited 4 Sep 2008). Available from: https://www.svims.ca/council/Ramar1.htm (accessed 29 October 2024)
- Brinkmann, W. (1897) Vorarbeiniten zu einer Pilzflora Westfalen. Jahresbericht des Westfälischen Provinzial-Vereins für Wissenschaft und Kunst 25: 195–207.
- Corner, E.J.H. (1970) Supplement to “a monograph of Clavaria and allied genera”. Beihefte zur Nova Hedwigia 33: 87–92.
- Deng, P.T., Liu, W.H., Ge, Z.W. & Zhang, P. (2024) Three new ramarioid species of Phaeoclavulina (Gomphaceae, Gomphales) from China. MycoKeys 108: 1–14. https://doi.org/10.3897/mycokeys.108.128716
- Elkhateeb, W., Elnahas, M., Lu, W.H., Galappaththi, M.C.A. & Daba, G.M. (2021) The coral mushrooms Ramaria and Clavaria. Studies in Fungi 6 (1): 495–506. https://doi.org/10.5943/sif/6/1/39
- Exeter, R.L., Norvell, L. & Cazares, E. (2006) Ramaria of the Pacific Northwestern United States. Bureau of Land Management, Salem, Oregon, 157 pp.
- Franchi, P. & Marchetti, M. (2018) Nomenclatural novelties. Index Fungorum 373: 1–1.
- Franchi, P. & Marchetti, M. (2019) Nuove combinazioni nel Genere Phaeoclavulina. Rivista di Micologia 62: 19–40.
- Franchi, P. & Marchetti, M. (2020) Nomenclatural novelties. Index Fungorum 457: 1–7.
- Gardes, M. & Bruns, T.D. (1993) ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Molecular Ecology 2 (2): 113–118.
- Giachini, A.J. (2004) Systematic, Phylogeny, and Ecology of Gomphus sensu lato. Oregon State University. Corvallis.
- Giachini, A.J., Hosaka, K., Nouhra, E., Spatafora, J. & Trappe, J.M. (2010) Phylogenetic relationships of the Gomphales based on nuc-25S-rDNA, mit12S-rDNA, and mit-atp6-DNA combined sequences. Fungal Biology 114: 224–234. https://doi.org/10.1016/j. funbio.2010.01.002
- Giachini, A.J. & Castellano, M.A. (2011) A new taxonomic classification for species in Gomphus sensu lato. Mycotaxon 115 (1): 183–201. https://doi.org/10.5248/115.183
- Giachini, A.J., Castellano, M.A. & Cázares, É. (2019) Systematics of the Gomphales: The Genus Phaeoclavulina Brinkmann. Mycotaxon.
- Glez-Peña, D., Gómez-Blanco, D., Reboiro-Jato, M., Fdez-Riverola, F., Posada, D. (2010) ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Research 38 (2): W14–W18. https://doi.org/10.1093/nar/gkq321
- González-Ávila, P., Torres-Miranda, A., Villegas-Ríos, M. & Luna-Vega, I. (2013) Species diversity and ecological patterns of Phaeoclavulina species in Mexico with implications for conservation. North American Fungi 8 (16): 1–32.
- González-Vila, A., Martínez-González, R., Espinosa, D. & Estrada-Torres, A. (2020) Phaeoclavulina liliputiana sp. nov. (Gomphaceae, Gomphales) a new endemic species from Tlaxcala, Mexico. Phytotaxa 470 (2): 155–164. https://doi.org/10.11646/phytotaxa.470.2.4
- Hibbett, D.S., Binder, M., Bischoff, J.F., Blackwell, M., Cannon, P.F., Eriksson, O.E., Huhndorf, S., James, T., Kirk, P.M., Lucking, R., Lumbsch, H.T., Lutzoni, F., Matheny, B.P., Mclaughlin, D.J., Powell, M.J., Redhead, S., Schoch, C.L., Spatafora, J.W., Stalpers, J.A., Vilgalys, R., Aime, C.M., Aptroot, A., Bauer, R., Begerow, D., Benny, G.L., Castlebury, L.A., Crous, P.W., Dai, Y.C., Gams, W., Geiser, D.M., Griffith, G.W., Gueidan, C., Hawksworth, D.L., Hestmark, G., Hosaka, K., Humber, R.A., Hyde, K.D., Ironside, J.E., Kõljalg, U., Kurtzman, C.P., Larsson, K.H., Lichtwardt, R., Longcore, J., Miadlikowska, J., Miller, A., Moncalvo, J.M., Standridge, S.M., Oberwinkler, F., Parmasto, E., Reeb, V., Rogers, J.D., Roux, C., Ryvarden, L., Sampaio, J.P., Schüßler, A., Sugiyama, J., Thorn, G.R., Tibell, L., Untereiner, W.A., Walker, C., Wang, Z., Weir, A., Weiss, M., White, M.M., Winka, K., Yao, Y.J. & Zhang, N. (2007) A higher-level phylogenetic classification of the fungi. Mycological Research 111 (5): 509–547. https://doi.org/10.1016/j.mycres.2007.03.004
- Hosaka, K., Bates, S.T., Beever, R.E., Castellano, M.A., Colgan, III. W., Dominguez, L.S., Nouhra, E.R., Geml, J., Giachini, A.J., Kenney, S.R., Simpson, N.B., Spatafora, J.W. & Trappe, J.M. (2006) Molecular phylogenetics of the gomphoid-phalloid fungi with an establishment of the new subclass Phallomycetidae and two new orders. Mycologia 98 (6): 949–959. https://doi.org/10.1080/15572536.2006.11832624
- Hu, Y., Karunarathna, S.C., Li, H., Galappaththi, M.C., Zhao, C.L., Kakumyan, P. & Mortimer, P.E. (2022) The impact of drying temperature on basidiospore size. Diversity 14 (4): 239. https://doi.org/10.3390/d14040239
- Katoh, K. & Standley, D.M. (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32 (13): 1933–1942. https:// doi.org/10.1093/bioinformatics/btw108
- Katoh, K., Rozewicki, J. & Yamada, K. (2019) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20 (4): 1160–1166. https://doi.org/10.1093/bib/bbx108
- Kornerup, A. & Wanscher, J.H. (1978) Methuen handbook of colour (3rd edn). Eyre Methuen, London, 252 pp.
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. In: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the campus and beyond. Association for Computing Machinery, New York, pp. 1–8. [Article No.: 39] https://doi.org/10.1145/2335755.2335836
- Li, J.Z. (2008) Study on wild edible fungi species diversity from Hunan. Life Science Research 12 (4): 314–321.
- Liu, W.H., Yan, J., Deng, P.T., Qin, W.Q. & Zhang, P. (2022) Two new species of Phaeoclavulina (Gomphaceae, Gomphales) from Hunan Province, China. Phytotaxa 561 (1): 27–40. https://doi.org/10.11646/phytotaxa.561.1.3
- Lu, W.H., Nutaratat, P., Kumla, J., Tibpromma, S., Elgorban, A.M., Karunarathna, S.C. & Suwannarach, N. (2024) Morphological and molecular identification of two new Marasmiellus species (Omphalotaceae, Agaricales) from Thailand. MycoKeys 109: 31–48. https://doi.org/10.3897/mycokeys.109.129791
- Nasim, G., Ali, M. & Shabbir, A. (2008) A study of genus Ramaria from Ayubia National Park, Pakistan. Mycopathology 6 (1–2): 43–46.
- Nylander, J.A.A. (2004) MrModeltest v2.0. Program distributed by the author, Evolutionary Biology Centre, Uppsala, Sweden.
- Petersen, R.H. (1981) Ramaria subgenus Echinoramaria. Mycologica 79: 1–261.
- Rambaut, A. & Drummond, A.J. (2012) FigTree version 1.4. 0. Available from: http://tree.bio.ed.ac.uk/software/figtree (accessed 29 October 2024)
- Rathnayaka, A.R., Tennakoon, D.S., Jones, G.E., Wanasinghe, D.N., Bhat, D.J., Priyashantha, A.H., Stephenson, S.L., Tibpromma, S. & Karunarathna, S.C. (2024) Significance of precise documentation of hosts and geospatial data of fungal collections, with an emphasis on plant-associated fungi. New Zealand Journal of Botany. [28 pp.] https://doi.org/10.1080/0028825X.2024.2381734
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542.
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172 (8): 4238–4246.
- Villegas, M., Cifuentes, J. & Estrada-Torres, A.E. (2005) Sporal characters in Gomphales and their significance for phylogenetics. Fungal Diversit 18: 157–175. https://doi.org/10.1016/j.femsyr.2005.01.001
- Stamatakis, A., Ludwig, T. & Meier, H. (2005) RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21 (4): 456–463. https://doi.org/10.1093/bioinformatics/bti191
- Stamatakis, A. (2006) RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 (21): 2688–2690. https://doi.org/10.1093/bioinformatics/btl446
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9): 1312–1313. https://doi.org/10.1093/bioinformatics/btu033.
- Teng, S.C. (1963) Fungi of China. Science Press. Beijing; pp. 1–808.
- White, T.J., Bruns, T., Lee, S., Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (ed.) PCR Protocols: A guide to methods and applications. San Diego, CA: Academic Press, New York, pp. 315–322.
- Zhang, P., Yang, Z.L. & Ge, Z.W. (2005) Two new species of Ramaria from southwestern China. Mycotaxon 94: 235–240.
- Zhou, H.M., Zhang, X.C., Li, J.T., Wu, F. & Zhao, C.L. (2024) Morphological characteristics and phylogenetic analyses revealed four new wood inhabiting fungi (Agaricomycetes, Basidiomycota) in Xizang Autonomous Region, China. MycoKeys 106: 20–224. https://doi.org/10.3897/mycokeys.106.125831