Abstract
The genus Bellevalia within the Asparagaceae family comprises 76 species distributed across various biogeographical regions. Despite previous taxonomic studies, there remains a gap in understanding the phylogenetic relationships among Bellevalia species.
This research was carried out in a situation where many species of this family do not have a special position in the classification, or their position has changed due to the use of different morphological characteristics.
In Feinbrun’s monograph in 1939, he used morphological traits such as the shape of the inflorescence, the direction of the peduncle, the color of the peduncle, the ratio of the length of the leaf to the flowerless stem, to separate the sections, sub-sections and species, and the interference of the traits can be seen in the separation of the arrays. On the other hand, it is difficult to refer to the color of fresh flowers in most of the herbarium samples because in these samples, the color of the flower cover gradually turns from yellow-brown to gray and it is difficult to identify them according to the color of dried flowers. It can be stated that due to the overlap between the morphological characteristics and the tendency of flower color to change in dry specimens, the features used by Feinbrun are not informative enough to recognize the circumscription of the sections.
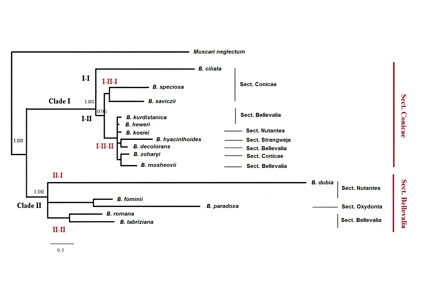
The objective is to resolve taxonomic ambiguities and enhance species identification accuracy. Freshly collected samples and herbarium specimens from 15 Bellevalia species were analyzed. Fifteen morphological characteristics, the most important of which are the flower, leaf margin cilia, Inflorescence shape and Capsule shape were recorded. The results identified two major clades in this genus and provide new phylogenetic insights. This is while the previous classifications of this genus, which were based only on morphological characteristics, included 4 to 6 sections. Key numerical outcomes include the clear separation of species, and enhancing taxonomic resolution. This study underscores the significance of integrating morphological and molecular data, presenting a robust framework for future phylogenetic studies within Bellevalia and related taxa.
References
- Al-Ali, H. (2016) The evolution of drug discovery: from phenotypes to targets, and back. MedChemComm 7 (5): 788–798.
- APG IV. (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Botanical Journal of Linnaean Society 181: 1–20.
- Astuti, G., Brullo, S., Domina, G., El Mokni, R., Giordani, T. & Peruzzi, L. (2017) Phylogenetic relationships among tetraploid species of Bellevalia (Asparagaceae) endemic to south-central Mediteranean. Plant Biosystems 151 (6): 1120–1128.
- Baker, J.G. (1871) A revision of genera and species of herbaceous capsular gamophyllous Liliaceae. Botanical Journal of the Linnean Society 11: 349–436.
- Boissier, E. (1884) Diagnosis plantarum orientales novarum. Bulletin de l‘Herbier Boissier, ser. 1 5: 300–308. [Geneva]
- Borzatti von Loewensten, A., Giordani, T., Astuti, G., Andreucci, A. & Peruzzi, L. (2013) Phylogenetic relationships of Italian Bellevalia species (Asparagaceae), inferred from morphology, karyology and molecular systematic. Plant Biosystems 147 (3): 776–787.
- Doyle, J.J. & Doyle, J.L. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
- Feinbrun, N. (1986) Flora of Palestine. The Israel academy of science & Humanity. Jerusalem 4: 84–104.
- Feinbrun, N. (1940) A monographic study of Bellevalia Lapeyr. Palestine journal of botany Jerusalem, 1: 336–340.
- Govaerts, R. (2021) World checklist of Asparagaceae. Available from: http://apps.kew.org/wcsp (accessed 23 April 2025)
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analyses program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
- Jafari, A., Vaezi, J., Forghanifard, M.M., Forest, F. & Manning, J.C. (2021) A revised infrageneric classification of Bellevalia Lapeyr. (Asparagaceae: Scilloideae) based on molecular analysis. Phytotaxa 525: 70–84. https://doi.org/10.11646/phytotaxa.525.1.9
- Jalali, L., Jafari, A., Vaezi, J. & Karimi, E. (2022) A simple and efficient DNA extraction protocol for old herbarium leaves of Bellevalia Lapeyr. (Asparagaceae, Scilloideae). Nova Biologica Reperta 9 (2): 124–131.
- Lapeyrouse, P.P. (1808) Bellevalia Lapeyr. Nouveau genre de plante de la famille des Liliacées. Journal de Physique, de Chimie, d’Historie Naturelle et des Arts 67: 425–427.
- Losinskaja, A.S.L. (1935) Bellevalia Lapeyr. In: Komarov, V.L. (Ed.) Flora of USSR 4. Akademiya Nauk SSSR, Leningrad, pp. 303–311.
- Manning, C., Forest, F., Devey, D.S., Fay, M. & Goldblatt, P. (2009) A molecular phylogeny and a revised classification of Ornithogaloideae (Hyacinthaceae) based on an analysis of four plastid DNA regions. Taxon 58 (1): 1–107. https://doi.org/10.1002/tax.581011
- Müller, K. (2005) SeqState: primer design and sequence statistics for phylogenetic DNA datasets. Applied bioinformatics 4: 65–69.
- Nylander, J.A.A. (2004) MrModeltest v2. Program distributed by the author, Evolutionary Biology Centre, Uppsala University. Available from: http://www.abc.se/~nylander/ (accessed 23 April 2025)
- Page, R.D.M. (2001) TreeView (Win32). Version 1.6.6. [http://taxonomy.zoology.gla.ac.uk/rod/rod.html]
- Persson, K. & Wendelbo, P. (1979) The taxonomic position of Bellevalia tabriziana (Liliaceae). Botaniska Notiser 132: 197–200.
- Pfosser, M. & Speta, F. (1999) Phylogenetic of Hyacinthaceae based on plastid DNA sequences. Annals of the Missouri Botanical Garden 86 (4): 852–875. https://doi.org/10.2307/2666172
- Posada, D. & Buckley, T.R. (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic biology 53 (5): 793–808.
- Rambaut, A. & Drummond, A.J. (2007) Tracer Version 1.5: MCMC trace analysis tool.
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes. 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Simmons, M.P. & Ochoterena, H. (2000) Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49: 369–381.
- Speta, F. (1998) Hyacinthaceae. In: Kubitzki, K. (ed.) The families and genera of vascular plants. III. Flowering plants. Monocotyledons. Liliaceae (except Orchidaceae). Springer, Berlin, pp. 261–285.
- Swofford, D.L. (2002) PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sinauer, Sunderland, MA.
- Thompson, P.G., Hong, L.L., Ukoskit, K. & Zhu, S. (1994) Genetic linkage of randomly amplified polymorphic DNA (RAPD) markers in sweet potato. Journal of the American Society for Horticultural Science 122: 79–82.
- Trifonova, A.A., Filyushin, M.A., E.Z. Kochieva, E.Z. & Kudryavtsev, A.M. (2016) Analysis of the ITS1/ITS2 Nuclear pacers and the secondary structure of 5.8S rRNA gene in endemic species Bellevalia sarmatica (Pall. ex Georgi) Woronow and Related Species of the Subfamily Scilloideae. Russian Journal of Genetics 52 (5): 530–534.
- Vaezi, J., Arjmandi, A.A. & Sharghi, H.R. (2019) Origin of Rosa × binaloudensis (Rosaceae), a new natural hybrid species from Iran. Phytotaxa 411 (1): 23–38. https://doi.org/10.11646/phytotaxa.411.1.2
- Vaezi, J. & Brouillet, L. (2022) Origin of Symphyotrichum anticostense (Asteraceae, Astereae), an endemic, high polyploid species of the Gulf of St. Lawrence region, based on morphological and nrDNA evidence. Botany 100 (7): 551–571.
- Wendelbo, P. (1980) Notes on Hyacinthus and Bellevalia (Liliaceae) in Turkey and Iran. Notes of the Royal Botanical Garden of Edinburgh 38 (3): 423–434.