Abstract
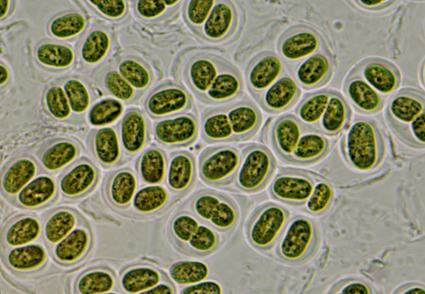
Cyanobacteria, with strong adaptability, occupied an important position on the earth all the time. In this study, the strain of coccoid cyanobacteria (CHAB 5847) was isolated and purified from Guizhou Province, China. The phylogenetic and taxonomic characteristics of CHAB 5847 were studied using polyphasic approach combining morphological, ecological and molecular data. Based on phylogenetic analysis of 16S rRNA gene sequence, the studied strain occupied a unique position, and clustered in Gloeothece clade. The 16S rRNA gene sequences of two clones of the studied strain shared 93.1%–98.1% similarity to other Gloeothece species, which supports well it to be a new member of Gloeothece genus. Furthermore, the unique pattern of D1-D1′, Box-B and V3 helix of the 16S–23S rRNA internal transcribed spacer (ITS) secondary structure revealed that the strain represented a novel species. These results support the establishment of a new species of Gloeothece genus with the name G. guizhouensis sp. nov.
References
- Bafford, R.A., Seagull, R.W., Chung, S.-Y. & Millie, D.F. (1993) Intracellular localization of the taste/odor metabolite 2-methylisoborneol in Oscillatoria limosa (cyanophyta). Journal of Phycology 29 (1): 91–95. https://doi.org/10.1111/j.1529-8817.1993.tb00285.x
- Boyer, S.L., Johansen, J.R., Flechtner, V.R. & Howard, G.L. (2002) Phylogeny and Genetic Variance in Terrestrial Microcoleus (Cyanophyceae) Species Based on Sequence Analysis of the 16S rRNA Gene and Associated 16S–23S ITS Region. Journal of Phycology 38 (6): 1222–1235. https://doi.org/10.1046/j.1529-8817.2002.01168.x
- Cai F., Yu, G., Zhang, K., Chen, Y., Li, Q., Yang, Y., Xie, J., Wang, Y. & Li, R. (2017) Geosmin production and polyphasic characterization of Oscillatoria limosa Agardh ex Gomont isolated from the open canal of a large drinking water system in Tianjin City, China. Harmful Algae 69: 28–37. https://doi.org/10.1016/j.hal.2017.09.006
- Cai, F., Yu, G., Liu, Y., Sun, Y. & Li, R. (2021) Description of two new species of Nostoc from China based on the polyphasic approach. Fottea 21 (2): 259–271. https://doi.org/10.5507/fot.2021.011
- Casamatta, D.A., Johansen, J.R., Vis, M.L. & Broadwater, S.T. (2005) Molecular and ultrastructural characterization of ten polar and nearpolar strains within the Oscillatoriales (Cyanobacteria). Journal of Phycology 41: 421–438. https://doi.org/10.1111/j.1529-8817.2005.04062.x
- Cassier-Chauvat, C. & Chauvat, F. (2014) Responses to oxidative and heavy metal stresses in cyanobacteria: recent advances. International journal of molecular sciences 16 (1): 871–886. https://doi.org/10.3390/ijms16010871
- Chen, W., Li, S., Xu, Y., Geng, R., Song, G. & Ma, P. (2023) Introducing Cyanodorina gen. nov. And Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach. Diversity 15 (3): 329. https://doi.org/10.3390/d15030329
- Debnath, M., Singh, T. & Bhadury, P. (2017) New records of Cyanobacterial morphotypes with Leptolyngbya indica sp. nov. from terrestrial biofilms of the Lower Gangetic Plain, India. Phytotaxa 316 (2): 101–120. https://doi.org/10.11646/phytotaxa.316.2.1
- Edwards, U., Rogall, T., Blöcker, H., Emde, M. & Böttger, E.C. (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research 17 (19): 7843–7853. https://doi.org/10.1093/nar/17.19.7843
- Engene, N., Cameron, C.R. & Gerwick, W.H. (2010) 16S rRNA gene heterogeneity in the filamentous marine cyanobacterial genus Lyngbya. Journal of Phycology 46 (3): 591–601. https://doi.org/10.1111/j.1529-8817.2010.00840.x
- Erwin, P.M. & Thacker, R.W. (2008) Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge hosts. Molecular Ecology 17: 2937–2947. https://doi.org/10.1111/j.1365-294X.2008.03808.x
- Genuário, D.B., Vaz, M.G.M.V., Hentschke, G.S., Sant’Anna, C.L. & Fiore, M.F. (2015) Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. International journal of systematic and evolutionary microbiology 65 (Pt 2): 663–675. https://doi.org/10.1099/ijs.0.070078-0
- Hentschke G.S. & Sant'Anna C.L. (2015) Current trends and prospects for cyanobacterial taxonomy—are only cultured populations enough?. Algological Studies 147 (1): 3–6. https://doi.org/10.1127/algol_stud/2014/0185
- Herrero, A. & Flore, E. (2008) The Cyanobacteria: Molecular Biology, Genomics and Evolution. Caister Academic Press, Poole, pp. xii+484. https://doi.org/10.21775/9781910190432
- Jasser, I., Panou, M., Khomutovska, N., Sandzewicz, M., Panteris, E., Niyatbekov, T., Łach, Ł., Kwiatowski, J., Kokociński, M. & Gkelis, S. (2022) Cyanobacteria in hot pursuit: Characterization of cyanobacteria strains, including novel taxa, isolated from geothermal habitats from different ecoregions of the world. Molecular phylogenetics and evolution 170: 107454. https://doi.org/10.1016/j.ympev.2022.107454
- Jiang, Y., Song, G., Pan, Q., Yang, Y. & Li, R. (2015) Identification of genes for anatoxin—a biosynthesis in Cuspidothrix issatschenkoi. Harmful Algae 46: 43–48. https://doi.org/10.1016/j.hal.2015.05.005
- Johansen, J.R., Kováčik, L., Casamatta, D.A., Fucikova, K. & Kastovsky, J. (2011) Utility of 16S–23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwigia 92: 283–302. https://doi.org/10.1127/0029-5035/2011/0092-0283
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/NMETH.4285
- Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30 (4): 772–780. https://doi.org/10.1093/molbev/mst010
- Khalifa, S.A.M., Shedid, E.S., Saied, E.M., Jassbi, A.R., Jamebozorgi, F.H., Rateb, M.E., Du, M., Abdel-Daim, M.M., Kai, G.Y., Al-Hammady, M.A.M., Xiao, J., Guo, Z. & El-Seedi, H.R. (2021) Cyanobacteria-From the Oceans to the Potential Biotechnological and Biomedical Applications. Marine drugs 19 (5): 241. https://doi.org/10.3390/md19050241
- Kim, M., Oh, H.S., Park, S.C. & Chun, J. (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology 64 (2): 346–351. https://doi.org/10.1099/ijs.0.059774-0
- Komárek, J., Cepák, V., Kaštovský, Jan. & Sulek, J. (2004) What are the cyanobacterial genera Cyanothece and Cyanobacterium? Contribution to the combined molecular and phenotype taxonomic evaluation of cyanobacterial diversity. Algological Studies 113 (1): 1–36. https://doi.org/10.1127/1864-1318/2004/0113-0001
- Komárek, J., Kaštovský, J., Mareš, J. & Johansen, J.R. (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86 (4): 295–335.
- Komárek, J. (2016) A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. European Journal Phycology 51 (3): 346–353. https://doi.org/10.1080/09670262.2016.1163738
- Komárek, J. (2020) Quo vadis, taxonomy of cyanobacteria (2019). Fottea 20 (1): 104–110. https://doi.org/10.5507/fot.2019.020
- Komárek, J. (2023) Taxonomic review of cyanobacteria 2021/2022 according to polyphasic evaluation. Fottea 23 (1): 141–148. https://doi.org/10.5507/fot.2022.017
- Konstantinou, D., Voultsiadou, E., Panteris, E. & Gkelis, S. (2021) Revealing new sponge-associated cyanobacterial diversity: Novel genera and species. Molecular phylogenetics and evolution 155: 106991. https://doi.org/10.1016/j.ympev.2020.106991
- Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33 (7): 1870–1874. https://doi.org/10.1093/molbev/msw054
- Lepère, C., Wilmotte, A. & Meyer, B. (2000) Molecular Diversity of Microcystis Strains (Cyanophyceae, Chroococcales) Based on 16S rDNA Sequences. Plant Ecology and Evolution 70 (2): 275–283. https://doi.org/10.2307/3668646
- Mai, T., Johansen, J.R., Pietrasiak, N., Bohunická, M. & Martin, M.P. (2018) Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 365 (1): 1–59. https://doi.org/10.11646/phytotaxa.365.1.1
- Mareš, J., Hauer, T., Komárek, J. & Compère, P. (2013a) (2195) Proposal to conserve the name Gloeothece (Cyanophyceae) with a conserved type. Taxon 62 (5): 1056–1056. https://doi.org/10.12705/625.24
- Mareš, J., Hrouzek, P., Kana, R., Ventura, S., Strunecky, O. & Komarek, J. (2013b) The Primitive Thylakoid-Less Cyanobacterium Gloeobacter Is a Common Rock-Dwelling Organism. Plos One 8 (6): e66323. https://doi.org/10.1371/journal.pone.0066323
- Mareš, J., Komárek, J., Compère, P. & Oren, A. (2013c) (2194) Proposal to conserve the name Gloeobacter violaceus against Aphanothece caldariorum, Gloeothece coerulea, and Gloeothece linearis (Cyanophyceae). Taxon 62 (5): 1055. https://doi.org/10.12705/625.23
- Mareš, J., Johansen, J.R., Hauer, T., Zima, J. Jr., Ventura, S., Cuzman, O., Tiribilli, B. & Kaštovský, J. (2019) Taxonomic resolution of the genus Cyanothece (Chroococcales, Cyanobacteria), with a treatment on Gloeothece and three new genera, Crocosphaera, Rippkaea, and Zehria. Journal of phycology 55 (3): 578–610. https://doi.org/10.1111/jpy.12853
- Martins, M.D., Machado-de-Lima, N.M. & Branco, L.H.Z. (2019) Polyphasic approach using multilocus analyses supports the establishment of the new aerophytic cyanobacterial genus Pycnacronema (Coleofasciculaceae, Oscillatoriales). Journal of Phycology 55 (1): 146–159. https://doi.org/10.1111/jpy.12805
- Miller, M.A., Schwartz, T., Pickett, B.E., He, S., Klem, E.B., Scheuermann, R.H., Passarotti, M., Kaufman, S. & O’Leary, M.A. (2015) A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evolutionary Bioinformatics 11: 43–48. https://doi.org/10.4137/EBO.S21501
- Nägeli, C.V. (1849) Gattungen einzelliger Algen: physiologisch und systematisch bearbeitet. Friedrich Schulthess, Zürich, 166 pp. https://doi.org/10.5962/bhl.title.6805
- Ohki, K., Kamiya, M., Honda, D., Kumazawa, S. & Ki, H.K. (2008) Morphological and phylogenetic studies on unicellular diazotrophic cyanobacteria (cyanophytes) isolated from the coastal waters around Singapore. Journal of Phycology 44 (1): 142–151. https://doi.org/10.1111/j.1529-8817.2007.00428.x
- Osorio-Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Kováčik, L., Martin, M.P. & Johensen, J.R. (2014) Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. European Journal of Phycology 49 (4): 450–470. https://doi.org/10.1080/09670262.2014.976843
- Pereira, S.B., Ow, S.Y., Barrios-Llerena, M.E., Wright, P.C., Moradas-Ferreira, P. & Tamagnini, P. (2011) iTRAQ-based quantitative proteomic analysis of Gloeothece sp. PCC 6909: Comparison with its sheathless mutant and adaptations to nitrate deficiency and sulfur limitation. Journal of proteomics 75 (1): 270–283. https://doi.org/10.1016/j.jprot.2011.09.007
- Pham, T.-L., Bui, M.H., Driscoll, M., Shimizu, K. & Motoo, U. (2021) First report of geosmin and 2-methylisoborneol (2-MIB) in Dolichospermum and Oscillatoria from Vietnam. Limnology 22 (1): 43–56. https://doi.org/10.1007/s10201-020-00630-2
- Pietrasiak, N., Mühlsteinová, R., Siegesmund, M. & Johansen, J.R. (2014) Phylogenetic placement of Symplocastrum (Phormidiaceae, Cyanobacteria) with descriptions of two new species: S. flechtnerae and S. torsivum. Phycologia 53 (6): 529–541. https://doi.org/10.2216/14-029.1
- Pöschl, U., Weber, B., Büdel, B., Burrows, S., Andreae, M.O. & Elbert, W. (2012) Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nature Geoscience 5 (7): 459–462. https://doi.org/10.1038/ngeo1486
- Prado, J., Hirai, R.Y., Shimizu, G.H. & Cantuária, P.C. (2017) The Nomenclature Section in Shenzhen (China) and the principal changes in the International Code of Nomenclature for Algae, Fungi, and Plants. Rodriguesia 68 (4): 1499–1503. https://doi.org/10.1590/2175-7860201768430
- Reuter, J.S. & Mathews, D.H. (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC bioinformatics 11 (1): 129. https://doi.org/10.1186/1471-2105-11-129
- Rippka, R. (1988) Isolation and purification of cyanobacteria. Methods in enzymology 167: 3–27. https://doi.org/10.1016/0076-6879(88)67004-2
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
- Sambrook, J. & Russell, D.W. (2001) Molecular cloning: A laboratory manual (Vol. 1, 3rd ed.). Cold Spring Harbor Laboratory Press, USA, 2344 pp.
- Schirrmeister, B.E., Antonelli, A. & Bagheri, H.C. (2011) The origin of multicellularity in cyanobacteria. BMC Evolutionary Biology 11 (1): 45. https://doi.org/10.1186/1471-2148-11-45
- Sciuto, K. & Moro, I. (2016) Detection of the new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S-23S ITS region. Molecular Phylogenetics and Evolution 105: 15–35. https://doi.org/10.1016/j.ympev.2016.08.010
- Shalygin, S., Shalygina, R.R., Redkina, V.V., Gargas, C.B. & Johansen, J.R. (2020) Description of Stenomitos kolaenensis and S. hiloensis sp. nov. (Leptolyngbyaceae, Cyanobacteria) with an emendation of the genus. Phytotaxa 440 (2): 108–128. https://doi.org/10.11646/phytotaxa.440.2.3
- Song, G., Xu, Y., Chen, W., Li, S., Ma, P. & Geng, R. (2023) Introducing Cyanodorina gen. nov. and Cyanodorina ovale sp. nov. (Microcystaceae, Chroococcales), a Novel Coccoid Cyanobacterium Isolated from Caohai Lake in China Based on a Polyphasic Approach. Diversity-Basel 15 (3): 1424–2818. https://doi.org/10.3390/d15030329
- Strunecký, O., Ivanova, A.P. & Mareš, J. (2023) An updated classification of cyanobacterial orders and families based on phylogenomic and polyphasic analysis. Journal of phycology 59 (1): 12–51. https://doi.org/10.1111/jpy.13304
- Svirčev, Z., Lalić, D., Bojadžija, S.G., Tokodi, N. & Drobac, B.D. (2019) Global geographical and historical overview of cyanotoxin distribution and cyanobacterial poisonings. Archives of Toxicology 93 (9): 2429–2481. https://doi.org/10.1007/s00204-019-02524-4
- Syed-Shabthar, S.M., Rosli, M.K., Mohd-Zin, N.A., Romaino, S.M., Fazly-Ann, Z.A., Mahani, M.C., Abas-Mazni, O., Zainuddin, R., Yaakop, S. & Md-Zain, B.M. (2013) The molecular phylogenetic signature of Bali cattle revealed by maternal and paternal markers. Molecular biology reports 40 (8): 5165–5176. https://doi.org/10.1007/s11033-013-2619-y
- Wang, Y., Cai, F., Jia, N. & Li, R. (2019) Description of a novel coccoid cyanobacterial genus and species Sinocapsa zengkensis gen. nov. sp. nov. (Sinocapsaceae, incertae sedis), with taxonomic notes on genera in Chroococcidiopsidales. Phytotaxa 409 (3): 146–160. https://doi.org/10.11646/phytotaxa.409.3.3
- Ward, D.M., Ferris, M.J., Nold, S.C. & Bateson, M.M. (1998) A Natural View of Microbial Biodiversity within Hot Spring Cyanobacterial Mat Communities. Microbiology and Molecular Biology Reviews 62 (4): 1353–1370. https://doi.org/10.1128/MMBR.62.4.1353-1370.1998
- Wilmotte, A. (1994) Molecular Evolution and Taxonomy of the Cyanobacteria. In: Bryant, D.A. (Ed.) The Molecular Biology of Cyanobacteria. Kluwer Academic Publisher, Dordrecht, pp. 1–25. https://doi.org/10.1007/978-94-011-0227-8_1
- Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.H., Whitman, W.B., Euzéby, J., Amann, R. & Rosselló-Móra, R. (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature reviews Microbiology 12 (9): 635–645. https://doi.org/10.1038/nrmicro3330
- Zhang, X., Sherman, D.M. & Sherman, L.A. (2014) The Uptake Hydrogenase in the Unicellular Diazotrophic Cyanobacterium Cyanothece sp. Strain PCC 7822 Protects Nitrogenase from Oxygen Toxicity. Journal of Bacteriology 196 (4): 840–849. https://doi.org/10.1128/jb.01248-13