Abstract
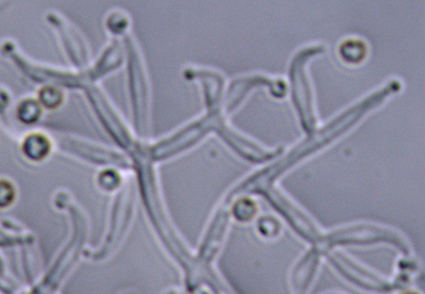
The orchid family is a natural resource pool, harboring numerous yet undiscovered species of endophytic fungi. However, few studies have investigated these endophytic fungi within Taiwan’s native orchids. In this study, two Trichoderma strains isolated from the roots of the native Taiwanese orchid, Dendrobium nakaharae, were identified as a new species, Trichoderma changiae. The phylogenetic analyses using three gene loci (ITS, tef1, and rpb2) revealed that the new species possesses distinctive sequences at these loci, clearly differentiating it from other species. Additionally, the analyses showed that T. changiae belongs to the Viride clade and is closely related to T. viride. The morphological and cultural characteristics of these isolates were observed, described, and illustrated in detail. Furthermore, the distinctions between this new species and its close relatives were compared, and the findings are presented herein.
References
- Cai, F. & Druzhinina, I.S. (2021) In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Diversity 107: 1–69. https://doi.org/10.1007/s13225-020-00464-4
- Carbone, I. & Kohn, L.M. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. https://doi.org/10.1080/00275514.1999.12061051
- Chand, K., Shah, S., Sharma, J., Paudel, M.R. & Pant, B. (2020) Isolation, characterization, and plant growth-promoting activities of endophytic fungi from a wild orchid Vanda cristata. Plant Signaling and Behavior 15: 1744294. https://doi.org/10.1080/15592324.2020.1744294
- Chen, K. & Zhuang, W.Y. (2017) Seven soil-inhabiting new species of the genus Trichoderma in the Viride clade. Phtotaxa 312: 28–46. https://doi.org/10.11646/phytotaxa.312.1.2
- Cheng, S.-F. (2012) The distribution and application of root fungal endophytes in Taiwan native orchids. [Doctoral dissertation, National Taiwan University] http://tdr.lib.ntu.edu.tw/jspui/handle/123456789/6461 [in Chinese]
- Chua, R.W., Song, K.P. & Ting, A.S.Y. (2022) Antimicrobial activities and phytochemical screening of endophytic fungi isolated from Cymbidium and Dendrobium orchids. South African Journal of Botany 151: 909–918. https://doi.org/10.1016/j.sajb.2022.11.015
- Chua, R.W. & Ting, A.S.Y. (2021) Fungal endophytes from orchidaceae: diversity and applications. In: Yadav, A.N. (Ed.) Recent trends in mycological research. Springer, Cham, pp. 391–426. https://doi.org/10.1007/978-3-030-68260-6_14
- Dutta, P., Deb, L. & Pandey, A.K. (2022) Trichoderma- from lab bench to field application: Looking back over 50 years. Frontiers in Agronomy 4: 932839. https://doi.org/10.3389/fagro.2022.932839
- Gardes, M. & Bruns, T.D. (1993) ITS primers with enhanced specificity for basidiomycetes — application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
- Ghazanfar, M.U., Raza, M., Raza, W. & Qamar, M.I. (2018) Trichoderma as potential biocontrol agent, its exploitation in agriculture: a review. Plant Protection 2: 109–135. Available from: https://api.semanticscholar.org/CorpusID:146065043
- Hawrot-Paw, M. & Stańczuk, A. (2023) From waste biomass to cellulosic ethanol by separate hydrolysis and fermentation (SHF) with Trichoderma viride. Sustainability 15: 168. https://doi.org/10.3390/su15010168
- Jaklitsch, W.M., Komon, M., Kubicek, C.P. & Druzhinina, I.S. (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97: 1365–1378. https://doi.org/10.1080/15572536.2006.11832743
- Jaklitsch, W.M., Samuels, G.J., Dodd, S.L., Lu, B.S. & Druzhinina, I.S. (2006) Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Studies in Mycology 56: 135–177. https://doi.org/10.3114/sim.2006.56.04
- Kornerup, A. & Wanscher, J.H. (1978) Methuen Handbook of Colour. Eyre Methuen, London, 252 pp.
- Li, T., Yang, W., Wu, S., Selosse, M.-A. & Gao, J. (2021) Progress and prospects of mycorrhizal fungal diversity in orchids. Frontiers in Plant Science 12: 646325. https://doi.org/10.3389/fpls.2021.646325
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Lodi, R.S., Peng, C., Dong, X., Deng, P. & Peng, L. (2023) Trichoderma hamatum and its benefits. Journal of Fungi 9: 994. https://doi.org/10.3390/jof9100994
- Ma, X., Nontachaiyapoom, S., Jayawardena, R.S., Yde, K.D., Gentekaki, E., Zhou, S., Qian, Y., Wen, T. & Kang, J. (2018) Endophytic Colletotrichum species from Dendrobium spp. in China and Northern Thailand. MycoKeys 43: 23–57. https://doi.org/10.3897/mycokeys.43.25081
- Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). New Orleans, LA, USA, pp. 1–8. https://doi.org/10.1145/2016741.2016785
- Nascimento Brito, V., Lana Alves, J., Sírio Araújo, K., de Souza Leite, T., Borges de Queiroz, C., Liparini Pereira, O. & de Queiroz, M.V. (2023) Endophytic Trichoderma species from rubber trees native to the Brazilian Amazon, including four new species. Frontiers in Microbiology 14: 1095199. https://doi.org/10.3389/fmicb.2023.1095199
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38: 3022–3027. https://doi.org/10.1093/molbev/msab120
- Tang, X., Yuan, Y. & Zhang, J. (2020) How climate change will alter the distribution of suitable Dendrobium habitats. Frontiers in Ecology and Evolution 8: 536339. https://doi.org/10.3389/fevo.2020.536339
- Wei, Y., Jin, J., Lin, Z., Lu, C., Gao, J., Li, J., Xie, Q., Zhu, W., Zhu, G. & Yang, F. (2023) Genome-wide identification, expression, and molecular characterization of the CONSTANS-like gene family in seven orchid species. International Journal of Molecular Sciences 24: 16825. https://doi.org/10.3390/ijms242316825
- White, T.J., Bruns, T.D., Lee, S.B. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols — a guide to methods and applications. Academic Press, San Diego, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Ye, C., Jing, T., Sha, Y., Mo, M. & Yu, Z. (2023) Two new Trichoderma species (Hypocreales, Hypocreaceae) isolated from decaying tubers of Gastrodiaelate. MycoKeys 99: 187–207. https://doi.org/10.3897/mycokeys.99.109404
- Zhang, L., Rammitsu, K., Tetsuka, K., Yukawa, T. & Ogura-Tsujita, Y. (2022) Dominant Dendrobium officinale mycorrhizal partners vary among habitats and strongly induce seed germination in vitro. Frontiers in Ecology and Evolution 10: 994641. https://doi.org/10.3389/fevo.2022.994641
- Zhang, G.Z., Yang, H.T., Zhang, X.J., Zhou, F.Y., Wu, X.Q., Xie, X.Y., Zhao, X.Y. & Zhou, H.Z. (2022) Five new species of Trichoderma from moist soils in China. MycoKeys 87: 133–157. https://doi.org/10.3897/mycokeys.87.76085
- Zhao, R., Mao, L.J. & Zhang, C.L. (2023) Three new species of Trichoderma (Hypocreales, Hypocreaceae) from soils in China. MycoKeys 97: 21–40. http://doi.org/10.3897/mycokeys.97.101635
- Zheng, H., Qiao, M., Lv, Y., Du, X., Zhang, K.Q. & Yu, Z. (2021) New species of Trichoderma isolated as endophytes and saprobes from southwest China. Journal of Fungi 7: 467. Available from: https://www.mdpi.com/2309-608X/7/6/467 (accessed 4 Jul. 2024) https://doi.org/10.3390/jof7060467