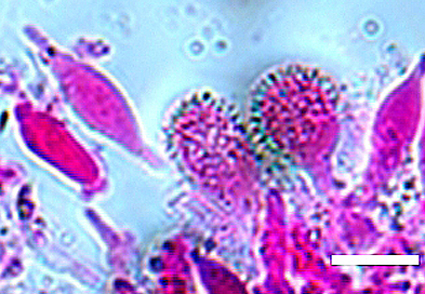
Abstract
We conducted a field investigation of marasmioid fungi in Jilin Province, China. Based on morphological and phylogenetic studies, seven specimens belonged to Cryptomarasmius (Physalacriaceae) were identified as three species. Phylogenetic analyses of ITS-nrLSU datasets conducted using Maximum Likelihood and Bayesian methods showed that C. changchunensis and C. aucubae are a new species and newly recorded species to China, respectively. Detailed descriptions and illustrations of the two species are provided in this study.
References
- Antonin, V. (2004) New species of marasmioid genera (Basidiomycetes, Tricholomataceae) from tropical Africa IV. Four new taxa of the genus Marasmius and one new combination. Mycotaxon 9 (2): 423–431.
- Antonin, V. & Noordeloos, M.E. (2010) A monograph of marasmioid and collybioid fungi in Europe. IHW Verlag, Eching, 480 pp.
- Bau, T. (2019) Mushroom in Jilin Agricultural University campus. Northeast Forestry University Press, Harbin, China, 270 pp.
- Bozok, F., Taskin, H., Büyükalaca, S., Dogan, H. & Assyov, B. (2018) Cryptomarasmius corbariensis (Physalacriaceae, Agaricales) in Turkey with first molecular data on the species from Eurasia. Nova Hedwigia 107 (1 & 2): 179–187. https://doi.org/10.1127/nova_hedwigia/2017/0462
- Corner, E.J.H. (1970) Supplement to “a monograph of Clavaria and allied genera”. Nova Hedwigia 33: 1–299.
- Desjardin, D.E. & Horak, E. (1997) Marasmius and Gloiocephala in the South Pacific region: Papua New Guinea, New Caledonia and New Zealand taxa. Bibliotheca Mycologica 168: 1−145.
- Desjardin, D.E., Retnowati, A. & Horak, E. (2000) Agaricales of Indonesia. 2. A preliminary monograph of Marasmius from Java and Bali. Sydowia 52: 92–193.
- Donk, M.A. (1964) A conspectus of the families of Aphyllophorales. Persoonia 3 (2): 199–324.
- Edler, D., Klein, J., Antonelli, A. & Silvestro, D. (2020) raxmlGUI 2.0 beta: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12 (2): 373–377. https://doi.org/10.1111/2041-210X.13512
- Jenkinson, T.S., Brian, A.P., Rainier, E.S. & Dennis, E.D. (2014) Cryptomarasmius gen. nov. established in the Physalacriaceae to accommodate members of Marasmius section Hygrometrici. Mycologia 106 (1): 86–94. https://doi.org/10.3852/11-309
- Kalyaanamoorthy, S., Minh, B., Wong, T.K.F., von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285
- Kiyashko, A.A., Malysheva, E.F., Antonin, V., Svetasheva, T.Y. & Bulakh, E.M. (2014) Fungi of the Russian Far East 2. New species and new records of Marasmius and Cryptomarasmius (Basidiomycota). Phytotaxa 186 (1): 1–28. https://doi.org/10.11646/phytotaxa.186.1.1
- Kornerup, A. & Wanscher, J.H. (1967) Methuen handbook of colour. Eyre Methuen, London, 252 pp.
- Kühner, R. (1933) Etudes sur le genre Marasmius. Botaniste 25: 57–115.
- Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33 (7): 1870–1874. https://doi.org/10.1093/molbev/msw054
- Lebel, T. & Catcheside, P. (2009) The truffle genus Cribbea (Physalacriaceae, Agaricales) in Australia. Australian Systematic Botany 22: 39–55. https://doi.org/10.1071/SB07041
- Moncalvo, J.M., Lutzoni, F.M., Rehner, S.A., Johnson, J. & Vilgalys, R. (2000) Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Systematic Biology 49 (2): 278–305. https://doi.org/10.1093/sysbio/49.2.278
- Moncalvo, J.-M., Vilgalys, R., Redhead, S.A., Johnson, J.E., James, T.Y., Aime, M.C., Hofstetter, V., Verduin, S.J.W., Larsson, E., Baroni, T.J., Thorn, R.G., Jacobsson, S., Clémençon, H. & Miller, O.K., Jr. (2002) One hundred and seventeen clades of euagarics. Molecular Phylogenetics and Evolution 23 (3): 357–400. https://doi.org/10.1016/S1055-7903(02)00027-1
- Moreno, G. & Raitviir, A. (1998) Marasmius celtibericus (Tricholomataceae, Agaricales) a new species from Spain. Persoonia—Molecular Phylogeny and Evolution of Fungi 16 (4): 541–544.
- Na, Q. & Bau, T. (2019) Recognition of Mycena sect. Amparoina sect. nov. (Mycenaceae, Agaricales), including four new species and revision of the limits of sect. Sacchariferae. MycoKeys 52: 103–124. https://doi.org/10.3897/mycokeys.52.34647
- Neda, H. & Doi, Y. (1998) Notes on agarics in Kyushu district. Memoirs of the National Science Museum Tokyo 31: 89–95.
- Pegler, D.N. (1982) Agaricoid and boletoid fungi (Basidiomycota) from Malawi and Zambia. Kew Bulletin 37 (2): 255–271. https://doi.org/10.2307/4109968
- Pegler, D.N. (1987) A revision of the Agaricales of Cuba. II: Species described by Earle and Murrill. Kew Bulletin 42 (4): 855–888. https://doi.org/10.2307/4109932
- Petch, T. (1947) A revision of Ceylon Marasmii. Transactions of the British Mycological Society 31 (1): 19–44. https://doi.org/10.1016/S0007-1536(47)80004-X
- Petrini, O., Petrini, L.E. & Horak, E. (1997) Taxonomic Monographs of Agaricales. II. Bibliotheca Mycologica 168: 1–152.
- Qin, J., Horak, E., Popa, F., Rexer, K.H., Kost, G., Li, F. & Yang, Z.L. (2018) Species diversity, distribution patterns, and substrate specificity of Strobilurus. Mycologia 110 (3): 584–604. https://doi.org/10.1080/00275514.2018.1463064
- Rambaut, A. (2019) FigTree v1. 4.3 2006–2016. Tree Figure Drawing Tool. Online publication. Institute of Evolutionary Biology, University of Edinburgh.
- Ronikier, M. & Ronikier, A. (2011) Rhizomarasmius epidryas (Physalacriaceae): phylogenetic placement of an arctic-alpine fungus with obligate saprobic affinity to Dryas spp. Mycologia 103 (5): 1124–1132. https://doi.org/10.3852/11-018
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Singer, R. (1945) New and interesting species of Basidiomycetes. Mycologia 37: 425–439. https://doi.org/10.1080/00275514.1945.12024003
- Singer, R. (1953) Type studies on Basidiomycetes. VI. Lilloa 26: 57–159.
- Singer, R. (1962) The Agaricales in modern taxonomy Ed.2. Weinheim: JCramer.
- Singer, R. (1986) The Agaricales in modern taxonomy Ed.4. Koenigstein: Koeltz Scientific Books.
- Singer, R. (1989) New taxa and new combinations of Agaricales (Diagnoses fungorum novorum Agaricalium IV). Fieldiana 21: 1–133. https://doi.org/10.5962/bhl.title.2537
- Song, H.B. & Bau, T. (2023) Conocybe Section Pilosellae in China: Reconciliation of Taxonomy and Phylogeny Reveals Seven New Species and a New Record. Journal of Fungi 9: 924. https://doi.org/ 10.3390/jof9090924
- Tan, Y.S., Desjardin, D.E., Perry, B.A., Vikineswary, S. & Noorlidah, A. (2009) Marasmius sensu stricto in peninsular Malaysia. Fungal Divers 37: 9–100.
- Vellinga, E.C. (1988) Glossary. In: Bas, C., Kuyper, T.H.W., Noordeloos, M.E. & Vellinga, E.C. (Eds.) Flora agaricina Neerlandica Volume 1. AABalkema, Rotterdam, Netherlands. 182 pp.
- Wang, G.S., Cai, Q., Hao, Y.J., Bau, T., Chen, Z.H., Li, M.X., David, N., Kraisitudomsook, N. & Yang, Z.L. (2023) Phylogenetic and taxonomic updates of Agaricales, with an emphasis on Tricholomopsis. Mycology. https://doi.org/10.1080/21501203.2023.2263031
- White, T.J., Bruns, T.D., Lee, S.B. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Zhang, D., Gao, F.L., Jakovlic, I., Zou, H., Zhang, J., Li, W.X. & Wang, G.T. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20 (1): 348–355. https://doi.org/10.1111/1755-0998.13096