Abstract
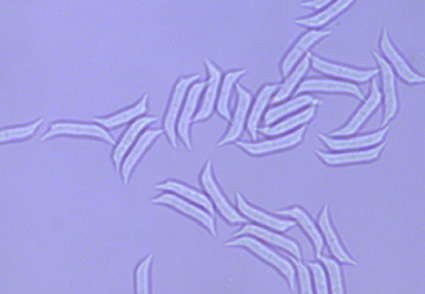
Arxiella longispora was isolated from forest litters in Northeast China. This new species is morphologically characterized by its shape and dimension of conidia and colonies on PDA and MEA. Multi-gene phylogenetic analysis based on a multi-gene dataset consisting of ITS, LSU, rpb2, and tef1 supported the placement of A. longispora in the genus Arxiella as a new species.
References
- Bills, G.F., Christensen, M., Powell, M. & Thorn, G. (2004) Saprobic soil fungi. Biodiversity of fungi: Inventory and monitoring methods: 271–302. https://doi.org/10.1016/b978-012509551-8/50016-7
- Crous, P.W., Wingfield, M.J., Schumacher, R.K., Summerell, B.A., Giraldo, A., Gené, J., Guarro, J., Wanasinghe, D.N., Hyde, K.D., Camporesi, E., Jones, E.B.G., Thambugala, K.M., Malysheva, E.F., Malysheva, V.F., Acharya, K., Álvarez, J., Alvarado, P., Assefa, A., Barnes, C.W., Bartlett, J.S., Blanchette, R.A., Burgess, T.I., Carlavilla, J.R., Coetzee, M.P., Damm, U., Decock, C.A., den Breeÿen, A., de Vries, B., Dutta, A.K., Holdom, D.G., Rooney-Latham, S., Manjón, J.L., Marincowitz, S., Mirabolfathy, M., Moreno, G., Nakashima, C., Papizadeh, M., Shahzadeh Fazeli, S.A., Amoozegar, M.A., Romberg, M.K., Shivas, R.G., Stalpers, J.A., Stielow, B., Stukely, M.J., Swart, W.J., Tan, Y.P., van der Bank, M., Wood, A.R., Zhang, Y. & Groenewald, J.Z. (2014) Fungal planet description sheets: 281–319. Persoonia: Molecular Phylogeny and Evolution of Fungi 33: 212–289. https://doi.org/10.3767/003158514X685680
- Endo, N., Ushijima, S., Nagasawa, E., Sugawara, R., Okuda, Y., Sotome, K., Nakagiri, A. & Maekawa, N. (2019) Taxonomic reconsideration of Tricholoma foliicola (Agaricales, Basidiomycota) based on basidiomata morphology, living culture characteristics, and phylogenetic analyses. Mycoscience 60 (6): 323–330. https://doi.org/10.1016/j.myc.2019.07.002
- Gao, S., Meng, W., Zhang, L., Yue, Q., Zheng, X. & Xu, L. (2021) Parametarhizium (Clavicipitaceae) gen. nov. with two new species as a potential biocontrol agent isolated from forest litters in Northeast China. Frontiers in microbiology 12: 131. https://doi.org/10.3389/fmicb.2021.627744
- Gönczöl, J. & Révay, Á. (2004) Fungal spores in rainwater: stemflow, throughfall and gutter conidial assemblages. Fungal Diversity 16: 67–86.
- Hernández-Restrepo, M., Bezerra, J.D.P., Tan, Y.P., Wiederhold, N., Crous, P.W., Guarro, J. & Gené, J. (2019) Re-evaluation of Mycoleptodiscus species and morphologically similar fungi. Persoonia: Molecular Phylogeny and Evolution of Fungi 42 (1): 205–227. https://doi.org/10.3767/persoonia.2019.42.08
- Jacob, M. (2000) Studies on Diversity, Ecology and Activity of the Microfungi Associated with Ficus benghalensis Linn. and Carissa congesta Wight from Goa State. Goa University, Taleigao, Goa, 346 pp.
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution 33 (7): 1870–1874. https://doi.org/10.1093/molbev/msw054
- Liang, J., Liu, B., Li, Z., Meng, W., Wang, Q. & Xu, L. (2019) Myxotrichum albicans, a new slowly-growing species isolated from forest litters in China. Mycoscience 60 (4): 232–236. https://doi.org/10.1016/j.myc.2019.03.002
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular biology and evolution 16 (12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Papendorf, M.C. (1967) Two new genera of soil fungi from South Africa. Transactions of the British Mycological Society 50 (1): 69–75. https://doi.org/10.1016/S0007-1536(67)80065-2
- Rehner, S.A. & Buckley, E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97 (1): 84–98. https://doi.org/10.1080/15572536.2006.11832842
- Ruscoe, Q. (1970) Arxiella lunata sp. nov. Transactions of the British Mycological Society 55 (3): 506–509. https://doi.org/10.1016/S0007-1536(70)80078-X
- Tennakoon, D.S., Kuo, C.H., Maharachchikumbura, S.S., Thambugala, K.M., Gentekaki, E., Phillips, A.J., Bhat, D.J., Wanasinghe, D.N., de Silva, N.I., Promputtha, I. & Hyde, K.D. (2021) Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Diversity 108 (1): 1–215. https://doi.org/10.1007/s13225-021-00474-w
- Vilgalys, R. & Sun BL. (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proceedings of the National Academy of Sciences 91 (10): 4599–4603. https://doi.org/10.1073/pnas.91.10.4599
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A.G.D., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, New York, pp. 315–322. https://doi.org/10.1016/b978-0-12-372180-8.50042-1