Abstract
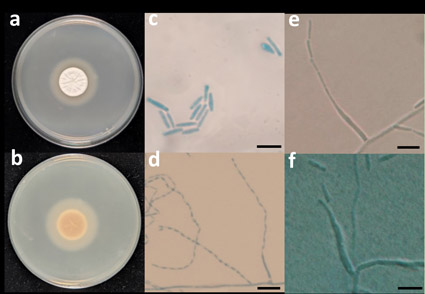
The investigation of fungal diversity in the rhizosphere of Camellia oleifera led to the isolation of two saprophytic fungal strains from the Xianyu Mountains, Anhui Province, China. Morphologically, these strains exhibited an irregular colony surface with divergent cracks, along with predominantly solitary slender phialides that produced narrow cylindrical conidia in chains. To further elucidate their taxonomic position, phylogenetic analyses were conducted using ITS and LSU rDNA region sequences, employing maximum-likelihood and Bayesian inference. The results unequivocally supported that these isolated strains belong to the genus Leptobacillium and represent a previously undescribed species. Consequently, based on their discernible morphological and phylogenetic characteristics in comparison to other Leptobacillium species, we propose a new fungal taxon, L. xianyushanense sp. nov.
References
- Chen, M.J., Han, Y.R., Hu, J.X., Liu, Y.J. & Huang, B. (2023) Tolypocladium rhizomatum sp. nov.: an endophytic species isolated from the rhizome of Polygonatum cyrtonema. Phytotaxa 606 (3): 201–210. https://doi.org/10.11646/phytotaxa.606.3.3
- Crous, P., Luangsa-Ard, J., Wingfield, M., Carnegie, A., Hernández-Restrepo, M., Lombard, L., Roux, J., Barreto, R., Baseia, I. & Cano-Lira, J. (2018) Fungal Planet description sheets: 785–867. Persoonia: Molecular Phylogeny and Evolution of Fungi 41: 238–417. https://doi.org/10.3767/persoonia.2018.41.12
- Daghino, S., Turci, F., Tomatis, M., Girlanda, M., Fubini, B. & Perotto, S. (2009) Weathering of chrysotile asbestos by the serpentine rock-inhabiting fungus Verticillium leptobactrum. FMS Microbiology Ecology 69 (1): 132–141. https://doi.org/10.1111/j.1574-6941.2009.00695.x
- Gams, W. & van Zaayen, A. (1982) Contribution to the taxonomy and pathogenicity of fungicolous Verticillium species. I. Taxonomy. Netherlands Journal of Plant Pathology 88: 57–78. https://doi.org/10.1007/BF01977339
- Gomes, A.A., Pinho, D.B., Cardeal, Z., Menezes, H.C., De Queiroz, M.V. & Pereira, O.L. (2018) Simplicillium coffeanum, a new endophytic species from Brazilian coffee plants, emitting antimicrobial volatiles. Phytotaxa 333 (2): 188–198. https://doi.org/10.11646/phytotaxa.333.2.2
- Hall, T. (2013) BioEdit v7.2.3. Biological sequence alignment editor for Win 95/98/NT/2K/XP7. California: Ibis Biosciences Carlsbad.
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20 (4): 1160–1166. https://doi.org/10.1093/bib/bbx108
- Leplat, J., Francois, A., Bousta, F. (2022) Leptobacillium cavernicola, a newly discovered fungal species isolated from several Paleolithic-decorated caves in France. Phytotaxa 571 (2): 186–196. https://doi.org/10.11646/phytotaxa.571.2.5
- Letunic, I. & Bork, P. (2019) Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Research 47 (1): 256–259. https://doi.org/10.1093/nar/gkz239
- Liu, F. & Cai, L. (2012) Morphological and molecular characterization of a novel species of Simplicillium from China. Cryptogamie Mycologie, 33 (2): 137–144. https://doi.org/10.7872/crym.v33.iss2.2012.137
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://github.com/nylander/MrModeltest2.
- Okane, I., Nonaka, K., Kurihara, Y., Abe, J.P. & Yamaoka, Y. (2020) A new species of Leptobacillim, L. symbioticum, isolated from mites and sori of soybean rust. Mycoscience 61 (4): 165–171. https://doi.org/10.1016/j.myc.2020.04.006
- Regaieg, H., Ciancio, A., Raouani, N.H. & Rosso, L. (2011) Detection and biocontrol potential of Verticillium leptobactrum parasitizing Meloidogyne spp. World Journal of Microbiology and Biotechnology 27 (7): 1615–1623. https://doi.org/10.1007/s11274-010-0615-0
- Spatafora, J.W., Volkmann-Kohlmeyer, B. & Kohlmeyer, J. (1998) Independent terrestrial origins of the Halosphaeriales (Marine Ascomycota). American Journal of Botany 85: 1569–1580. https://doi.org/10.2307/2446483
- Sun, J.Z., Ge, Q.Y., Zhu, Z.B., Zhang, X.L. & Liu, X.Z. (2019) Three dominating Hypocrealean fungi of the ‘white mold spots’ on acrylic varnish coatings of the murals in a Koguryo tomb in China. Phytotaxa 397 (3): 225–236. https://doi.org/10.11646/phytotaxa.397.3
- Trifinopoulos J, Nguyen, L.T, von Haeseler, A. & Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44: 232–235. https://doi.org/10.1093/nar/gkw256
- Vaidya, G., Lohman, D.J. & Meier, R. (2011) Sequence Matrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172 (8): 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- White, T.J., Bruns, T., Lee, S. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics.In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR protocols: a guide to methods and applications. Academic Press, New York, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Zare, R. & Gams, W. (2001) A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 73: 1–50. https://doi.org/10.1127/nova.hedwigia/71/2001/1
- Zare, R. & Gams, W. (2016) More white Verticillium-like anamorphs with erect conidiophores. Mycological Progress 15: 993–1030. https://doi.org/10.1007/s11557-016-1214-8
- Zare, R., Gams, W. & Culham, A. (2000) A revision of Verticillium sect. Prostrata. I. Phylogenetic studies using ITS sequences. Nova Hedwigia 71: 465–480. https://doi.org/10.1127/nova/71/2000/465