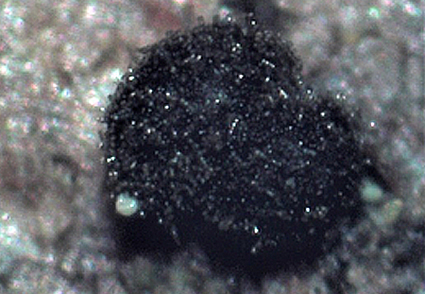
Abstract
A novel anamorphic ascomycetous fungus, Helminthosporium paraoligosporum (Pleosporales, Ascomycota), isolated from branches of linden trees is described and characterized. The species is morphologically distinct from other Helminthosporium spp. by shorter conidiophores and conidia, and culture characteristics. Phylogenetic analysis of a combined set of ITS and LSU sequence data also supports the establishment of a new species. H. paraoligosporum is clustered with H. caespitosum, H. oligosporum, and H. tiliae. The species in this clade have similar morphological characteristics and host affiliation. In particular, conidiophores of the species (except H. tiliae) can be reduced to elongated conidiogenous cells. H. caespitosum is associated with Betula spp., while the other species in this clade have been collected from Tilia spp. only.
References
- Alcorn, J.L. (1988) The taxonomy of “Helminthosporium” species. Annual Review of Phytopathology 26: 37–56. https://doi.org/10.1146/annurev.py.26.090188.000345
- Boonmee, S., Wanasinghe, D.N., Calabon, M.S., Huanraluek, N., Chandrasiri, S.K., Jones, E.B.G., Rossi, W., Leonardi, M., Singh, S.K., Rana, S., Singh, P.N., Maurya, D.K., Lagashetti, A.C., Choudhary, D., Dai, Y.C., Zhao, C.L., Mu, Y.H., Yuan, H.S., He, S.H., Phookamsak, R., Jiang, H.B., Martiìn, M.P., DuenÞas, M., Telleria, M.T., Kałucka, I.L., Jagodzinìski, A.M., Liimatainen, K., Pereira, D.S., Phillips, A.J.L., Suwannarach, N., Kumla, J., Khuna, S., Lumyong, S., Potter, T.B., Shivas, R.G., Sparks, A.H., Vaghefi, N., Abdel-Wahab, M.A., Abdel-Aziz, F.A., Li, G.J., Lin, W.F., Singh, U., Bhatt, R.P., Lee, H.B., Nguyen, T.T.T., Kirk, P.M., Dutta, A.K., Acharya, K., Sarma, V.V., Niranjan, M., Rajeshkumar, K.C., Ashtekar, N., Lad, S., Wijayawardene, N.N., Bhat, D.J., Xu, R.J., Wijesinghe, S.N., Shen, H.W., Luo, Z.L., Zhang, J.Y., Sysouphanthong, P., Thongklang, N., Bao, D.F., Aluthmuhandiram, J.V.S., Abdollahzadeh, J., Javadi, A., Dovana, F., Usman, M., Khalid, A.N., Dissanayake, A.J., Telagathoti, A., Probst, M., Peintner, U., Garrido-Benavent, I., Boìna, L., Mereìnyi, Z., Boros, L., Zoltaìn, B., Stielow, J.B., Jiang, N., Tian, C.M., Shams, E., Dehghanizadeh, F., Pordel, A., Javan-Nikkhah, M., Denchev, T.T., Denchev, C.M., Kemler, M., Begerow, D., Deng, C.Y., Harrower, E., Bozorov, T., Kholmuradova, T., Gafforov, Y., Abdurazakov, A., Xu, J.C., Mortimer, P.E., Ren, G.C., Jeewon, R., Maharachchikumbura, S.S.N., Phukhamsakda, C., Mapook, A. & Hyde, K.D. (2021) Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 111: 1–335. https://doi.org/10.1007/s13225-021-00489-3
- Chen, Y., Tian, W., Guo, Y., Madrid, H. & Maharachchikumbura, S.S.N. (2022) Synhelminthosporium gen. et sp. nov. and two new species of Helminthosporium (Massarinaceae, Pleosporales) from Sichuan Province, China. Journal of Fungi 8 (7): 712. https://doi.org/10.3390/jof8070712
- Crous, P.W., Wingfield, M.J., Lombard, L., Roets, F., Swart, W.J., Alvarado, P., Carnegie, A.J., Moreno, G., Luangsa-Ard, J., Thangavel, R., Alexandrova, A.V., Baseia, I.G., Bellanger, J.M., Bessette, A.E., Bessette, A.R., Delapeña-Lastra, S., García, D., Gené, J., Pham, T.H.G., Heykoop, M., Malysheva, E., Malysheva, V., Martín, M.P., Morozova, O.V., Noisripoom, W., Overton, B.E., Rea, A.E., Sewall, B.J., Smith, M.E., Smyth, C.W., Tasanathai, K., Visagie, C.M., Adamčík, S., Alves, A., Andrade, J.P., Aninat, M.J., Araújo, R.V.B., Bordallo, J.J., Boufleur, T., Baroncelli, R., Barreto, R.W., Bolin, J., Cabero, J., Caboň, M., Cafà, G., Caffot, M.L.H., Cai, L., Carlavilla, J.R., Chávez, R., Decastro, R.R.L., Delgat, L., Deschuyteneer, D., Dios, M.M., Domínguez, L.S., Evans, H.C., Eyssartier, G., Ferreira, B.W., Figueiredo, C.N., Liu, F., Fournier, J., Galli-Terasawa, L.V., Gil-Durán, C., Glienke, C., Gonçalves, M.F.M., Gryta, H., Guarro, J., Himaman, W., Hywel-Jones, N., Iturrieta-González, I., Ivanushkina, N.E., Jargeat, P., Khalid, A.N., Khan, J., Kiran, M., Kiss, L., Kochkina, G.A., Kolařík, M., Kubátová, A., Lodge, D.J., Loizides, M., Luque, D., Manjón, J.L., Marbach, P.A.S., Massolajr, N.S., Mata, M., Miller, A.N., Mongkolsamrit, S., Moreau, P.A., Morte, A., Mujic, A., Navarro-Ródenas, A., Németh, M.Z., Nóbrega, T.F., Nováková, A., Olariaga, I., Ozerskaya, S.M., Palma, M.A., Petters-Vandresen, D.A.L., Piontelli, E., Popov, E.S., Rodríguez, A., Requejo, Ó., Rodrigues, A.C.M., Rong, I.H., Roux, J., Seifert, K.A., Silva, B.D.B., Sklenář, F., Smith, J.A., Sousa, J.O., Souza, H.G., Desouza, J.T., Švec, K., Tanchaud, P., Tanney, J.B., Terasawa, F., Thanakitpipattana, D., Torres-Garcia, D., Vaca, I., Vaghefi, N., Vaniperen, A.L., Vasilenko, O.V., Verbeken, A., Yilmaz, N., Zamora, J.C., Zapata, M., Jurjevi, Ž. & Groenewald, J.Z. (2019) Fungal planet description sheets: 868–950. Persoonia 42: 291–473. https://doi.org/10.3767/persoonia.2019.42.11
- Darriba, D., Posada, D., Kozlov, A.M., Stamatakis, A., Morel, B. & Flouri, T. (2020) ModelTest–NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37: 291–294. https://doi.org/10.1093/molbev/msz189
- Errampalli, D., Saunders, J.M. & Holley, J.D. (2001) Emergence of silver scurf (Helminthosporium solani) as an economically important disease of potato. Plant Pathology 50: 141–153. https://doi.org/10.1046/j.1365-3059.2001.00555.x
- Hillis, D.M. & Bull, J.J. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. https://doi.org/10.1093/sysbio/42.2.182
- Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie ,E.H.C., Sarma, V.V., Boonmee, S., Lücking, R., Bhat, D.J., Liu, N.G., Tennakoon, D.S., Pem, D.2, Karunarathna, A., Jiang, S.H., Jones, E.B.G.18, Phillips, A.J.L., Manawasinghe, I.S., Tibpromma, S., Jayasiri, S.C., Sandamali, D.S., Jayawardena, R.S.,2, Wijayawardene, N.N., Ekanayaka, A.H., Jeewon, R., Lu, Y.Z., Dissanayake, A.J., Zeng, X.Y., Luo, Z.L., Tian, Q., Phukhamsakda, C., Thambugala, K.M., Dai, D.Q., Chethana, K.W.T., Samarakoon, M.C., Ertz, D., Bao, D.F., Doilom, M., Liu, J.K., Pérez-Ortega, S., Suija, A., Senwanna, C., Wijesinghe, S.N., Konta, S., Niranjan, M., Zhang, S.N., Ariyawansa, H.A., Jiang, H.B., Zhang, J.F., Norphanphoun, C., de Silva, N.I., Thiyagaraja, V., Zhang, H., Bezerra, J.D.P., Miranda-González, R., Aptroot, A., Kashiwadani, H., Harishchandra, D., Sérusiaux, E., Aluthmuhandiram, J.V.S., Abeywickrama, P.D., Devadatha, B., Wu, H.X., Moon, K.H., Gueidan, C., Schumm, F., Bundhun, D., Mapook, A., Monkai, J., Chomnunti, P., Suetrong, S., Chaiwan, N., Dayarathne, M.C., Yang, J., Rathnayaka, A.R., Bhunjun, C.S., Xu, J.C., Zheng, J.S., Liu, G., Feng, Y. & Xie, N. (2020) Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105: 17–318. https://doi.org/10.1007/s13225-020-00462-6
- Hu, Y-F., Liu, J-W., Xu, Z-H., Castañeda-Ruíz, R.F., Zhang, K. & Ma, J. (2023) Morphology and multigene phylogeny revealed three new species of Helminthosporium (Massarinaceae, Pleosporales) from China. Journal of Fungi 9 (2): 280. https://doi.org/10.3390/jof9020280
- Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Hughes, S.J. (1983) Helminthosporium oligosporum. In: Fungi Canadenses. no. 245.
- Konta, S., Hyde, K.D., Karunarathna, S.C., Mapook, A., Senwanna, C., Dauner, L.A.P., Nanayakkara, C.M., Xu, J., Tibpromma, S. & Lumyong, S. (2021) Multi-gene phylogeny and morphology reveal Haplohelminthosporium gen. nov. and Helminthosporiella gen. nov. associated with palms in Thailand and a checklist for Helminthosporium reported worldwide. Life (Basel) 11 (5): 454. https://doi.org/10.3390/life11050454
- Link, J.H.F. (1809) Observationes in ordines plantarum naturales. Dissertatio 1ma. Magazin der Gesellschaft Naturforschender Freunde zu Berlin 3: 3–42.
- Manzar, N., Kashyap, A.S., Maurya, A., Rajawat, M.V.S., Sharma, P.K., Srivastava, A.K., Roy, M., Saxena, A.K. & Singh, H.V. (2022) Multi-gene phylogenetic approach for identification and diversity analysis of Bipolaris maydis and Curvularia lunata isolates causing foliar blight of Zea mays. Journal of Fungi 8 (8): 802. https://doi.org/10.3390/jof8080802
- Marin-Felix, Y., Hernández-Restrepo, M. & Crous, P.W. (2020) Multi-locus phylogeny of the genus Curvularia and description of ten new species. Mycological Progress 19: 559–588. https://doi.org/10.1007/s11557-020-01576-6
- Phookamsak, R., Norphanphoun, C., Tanaka, K., Dai, D.-Q., Luo, Z.-L., Liu, J.-K., Su, H.-Y., Bhat, D.J., Bahkali, A.H., Mortimer, P.E., Xu, J.-C. & Hyde, K.D. (2015) Towards a natural classification of Astrosphaeriella-like species; Introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Diversity 74: 143–197. https://doi.org/10.1007/s13225-015-0352-7
- Posada, D. & Buckley, T. (2004) Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793–808. https://doi.org/10.1080/10635150490522304
- Rambaut, A. (2018) FigTree, version 1.4.4. Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK.
- Stamatakis, S. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Subramanian, C.V. & Sekar, G. (1987) The bitunicate ascomycetes and their anamorphs. Kavaka 15: 87–98.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. https://doi.org/10.1093/molbev/mst197
- Tanaka, K., Hirayama, K., Yonezawa, H., Sato, G., Toriyabe, A., Kudo, H., Hashimoto, A., Matsumura, M., Harada, Y., Kurihara, Y., Shirouzu, T. & Hosoya, T. (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82: 75–136. http://doi.org/10.1016/j.simyco.2015.10.002
- Thompson, J.D., Higgins, D.G. & Gibson, T.J. (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. https://doi.org/10.1093/nar/22.22.4673
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- White, T.J., Bruns, T.D., Lee, S. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR-protocols and applications- A laboratory manual. In: PCR Protocols: A Guide to Methods and Applications. pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Voglmayr, H. & Jaklitsch, W.M. (2017) Corynespora, Exosporium and Helminthosporium revisited – New species and generic reclassification. Studies in Mycology 87: 43–76. https://doi.org/10.1016/j.simyco.2017.05.001
- Zhao, N., Luo, Z.L., Hyde, K.D., Su, H.Y., Bhat, D.J., Liu, J.K., Bao, D.F. & Hao, Y.E. (2018) Helminthosporium submersum sp. nov. (Massarinaceae) from submerged wood in north-western Yunnan Province, China. Phytotaxa 348 (4): 269–278. https://doi.org/10.11646/phytotaxa.348.4.3
- Zhu, D., Luo, Z.L., Baht, D.J., Mckenzie, E.H.C., Bahkali, A.H., Zhou, D.Q., Su, H.Y. & Hyde, K.D. (2016) Helminthosporium velutinum and H. aquaticum sp. nov. from aquatic habitats in Yunnan Province, China. Phytotaxa 253 (3): 179–190. https://doi.org/10.11646/phytotaxa.253.3.1