Abstract
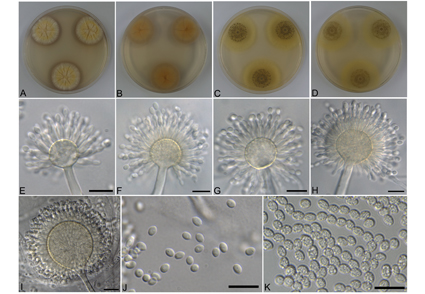
Aspergillus guangdongensis was included in the Aspergillus section Ochraceorosei. A notable feature of this section is that the majority of the described species are recognized only from the type strain. A. guangdongensis accommodates only a single strain, and the intraspecific variation within this species has not been realized. In investigating fungi from soil and stored plant seeds, two isolates, namely, BCRC 32544 and BCRC 32546, were identified as A. guangdongensis newly recorded from Taiwan. This is the first global record since its original description. To the best of our knowledge, all known species of section Ochraceorosei have not been previously reported in Taiwan. Our study also involved the phylogenetic analyses based on four loci genes (ITS, BenA, CaM, and RPB2), as well as a comparison of cultural and morphological characteristics between strains of A. guangdongensis and among closely related species, revealing obvious intraspecific variability. Aspergillus spp. produce a diverse array of important secondary metabolites, which have motivated great interests in scientific research groups. Therefore, a brief summary of research articles on the metabolites produced by the related species within the series Funiculosi under the Aspergillus section Ochraceorosei have been presented in this study.
References
- Bartoli, A. & Maggi, O. (1978) Four new species of Aspergillus from Ivory Coast soil. Transactions of the British Mycological Society 71: 383–394. https://doi.org/10.1016/S0007-1536(78)80064-3
- Cary, J.W., Ehrlich, K.C., Beltz, S.B., Harris-Coward, P. & Klich, M.A. (2009) Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia 101: 352–362. https://doi.org/10.3852/08-173
- Couteau, C. & Coiffard, L. (2016) Overview of skin whitening agents: drugs and cosmetic products. Cosmetics 3: 27. https://doi.org/10.3390/cosmetics3030027
- Frisvad, J.C., Skouboe, P. & Samson, R.A. (2005) Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Systematic and Applied Microbiology 28: 442–453. https://doi.org/10.1016/j.syapm.2005.02.012
- Gams, W., Christensen, M., Onions, A.H., Pitt, J.I. & Samson, R.A. (1986) Infrageneric Taxa of Aspergillus. In: Samson, R.A. & Pitt, J.I. (Eds.) Advances in Penicillium and Aspergillus Systematics. Springer, Boston, pp. 55–62. https://doi.org/10.1007/978-1-4757-1856-0_5
- Gardes, M. & Bruns, T.D. (1993) ITS primers with enhanced specificity for basidiomycetes — application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
- Glass, N.L. & Donaldson, G.C. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. https://journals.asm.org/doi/10.1128/aem.61.4.1323-1330.1995
- Han, L., Zheng, W., He, Z., Qian, S., Ma, X. & Kang, J. (2023) Endophytic fungus Biscogniauxia petrensis produces antibacterial substances. PeerJ 11: e15461. https://doi.org/10.7717/peerj.15461
- Houbraken, J., Kocsubé, S., Visagie, C.M., Yilmaz, N., Wang, X.C., Meijer, M., Kraak, B., Hubka, V., Samson, R.A. & Frisvad, J.C. (2020) Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 96: 141–153. https://doi.org/10.1016/j.simyco.2020.05.002
- Houbraken, J. & Samson, R.A. (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. https://doi.org/10.3114/sim.2011.70.01
- Kjærbølling, I., Vesth, T.C., Frisvad, J.C., Nybo, J.L., Theobald, S., Kuo, A., Bowyer, P., Matsuda, Y., Mondo, S., Lyhne, E.K., Kogle, M.E., Clum, A., Lipzen, A., Salamov, A., Ngan, C.Y., Daum, C., Chiniquy, J., Barry, K., LaButti, K., Haridas, S., Simmons, B.A., Magnuson, J.K., Mortensen, U.H., Larsen, T.O., Grigoriev, I.V., Baker, S.E. & Andersen, M.R. (2018) Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proceedings of the National Academy of Sciences 115: E753–E761. https://doi.org/10.1073/pnas.1715954115
- Kornerup, A. & Wanscher, J.H. (1978) Methuen Handbook of Colour. Eyre Methuen, London, 252 pp.
- Liu, Y.J., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1080/15572536.2008.11832477
- Moore, G.G., Mack, B.M. & Beltz, S.B. (2016) Draft genome sequences of two closely related aflatoxigenic Aspergillus species obtained from the Ivory Coast. Genome Biology and Evolution 8: 729–732. https://doi.org/10.1093/gbe/evv246
- Nakamura, M., Fukuyama, K., Tsukihara, T., Katsube, Y. & Hamasaki, T. (1983) Structure of funicin, antimicrobial substance from Aspergillus funiculosus, C17H18O5. Acta Crystallographica Section C 39: 268–270. https://doi.org/10.1107/S0108270183004357
- Nazir, M., Saleem, M., Tousif, M.I., Anwar, M.A., Surup, F., Ali, I., Wang, D., Mamadalieva, N.Z., Alshammari, E., Ashour, M.L., Ashour, A.M., Ahmed, I., Elizbit, Green, I.R. & Hussain, H. (2021) Meroterpenoids: a comprehensive update insight on structural diversity and biology. Biomolecules 11: 957. https://doi.org/ 10.3390/biom11070957
- Peterson, S.W. (2008) Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. https://doi.org/10.1080/15572536.2008.11832477
- Phasha, V., Senabe, J., Ndzotoyi, P., Okole, B., Fouche, G. & Chuturgoon, A. (2022) Review on the use of kojic acid—a skin-lightening ingredient. Cosmetics 9: 64. https://doi.org/10.3390/cosmetics9030064
- Raper, K.B. & Fennell, D.I. (1965) The Aspergillus sparsus group. In: Raper, K.B. & Fennell, D.I. (Eds.) The genus Aspergillus. Williams and Wilkins, Baltimore, pp. 431–441.
- Samson, R.A., Visagie, C.M., Houbraken, J., Hong, S.B., Hubka, V., Klaassen, C.H., Perrone, G., Seifert, K.A., Susca, A., Tanney, J.B., Varga, J., Kocsubé, S., Szigeti, G., Yaguchi, T. & Frisvad, J.C. (2014) Phylogeny, identification and nomenclature of the genus Aspergillus. Study in Mycolology 78: 141–173. https://doi.org/10.1016/j.simyco.2014.07.004
- Siddhardha, B., Murty, U.S., Narasimhulu, M. & Venkateswarlu, Y. (2010) Isolation, characterization and biological evaluation of secondary metabolite from Aspergillus funiculosus, C17H18O5. Indian Journal of Microbiology 50: 225–228. https://doi.org/10.1007/s12088-010-0044-7
- Smith, G. (1956) Some new species of soil moulds. Transactions of the British Mycological Society 39: 111–114 . https://doi.org/10.1016/S0007-1536(56)80059-4
- Sun, B., Luo, C., Bills, G.F., Li, J., Huang, P., Wang, L., Jiang, X. & Chen, A.J. (2022) Four new species of Aspergillus subgenus Nidulantes from China. Journal of Fungi 8: 1205. https://doi.org/10.3390/jof8111205
- Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38: 3022–3027. https://doi.org/10.1093/molbev/msab120
- White, T.J., Bruns, T.D., Lee, S.B. & Taylor J,W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols — a Guide to Methods and Applications. Academic Press, San Diego, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Winter, G. (1884) Familie. Perisporieae. In: Grunow, A., Hauck,F., Limpricht, G., Luerssen, Ch., Richter, P. & Winter, G. (Eds.) Dr. L. Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz, Edn 2. Verlag von Eduard Kummer, Leipzig, pp. 43–78. [https://www.biodiversitylibrary.org/item/16353#page/5/mode/1up]
- Yan, D. & Matsuda, Y. (2021) Genome mining-driven discovery of 5-methylorsellinate-derived meroterpenoids from Aspergillus funiculosus. Organic Letters 23: 3211–3215. https://doi.org/10.1021/acs.orglett.1c00951