Abstract
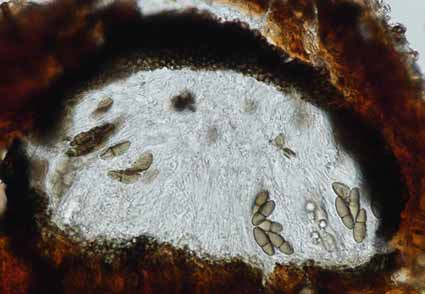
Fungal diversity is rapidly expanding, with new species being discovered worldwide. Northern Thailand is considered a hotspot for fungal diversity. In this study, we collected a saprobic microfungus from decaying wood in Chiang Mai Province, Thailand. Morphological examination and phylogenetic analyses of ITS, LSU, and SSU sequences using maximum likelihood and Bayesian inference placed a new isolate within Kirschsteiniothelia. Kirschsteiniothelia chiangmaiensis sp. nov. is characterised by 8-spored, cylindrical-clavate asci with a small ocular chamber and 1-septate ascospores that are ellipsoid to fusiform, with distinct guttules and a smooth wall surrounded by a mucilaginous sheath. Kirschsteiniothelia chiangmaiensis differs from the type by the smaller ascomata, wider asci, and presence of pluri-guttulate, and diverges from K. thujina by smaller ascomata, ascospore, and the presence of mucilaginous sheath. Based on morphology and phylogeny evidence, we introduce K. chiangmaiensis as a new species.
References
- Bao, D.F., Luo, Z.L., Liu, J.K., Bhat, D.J., Sarunya, N., Li, W.L., Su, H.Y. & Hyde, K.D. (2018) Lignicolous freshwater fungi in China III: Three new species and a new record of Kirschsteiniothelia from northwestern Yunnan Province. Mycosphere 9: 755–768. https://doi.org/10.5943/mycosphere/9/4/4
- Boonmee, S., Ko, T.W.K., Chukeatirote, E., Hyde, K.D., Chen, H., Cai, L., Mckenzie, E.H.C., Jones, E.B.G., Kodsueb, R. & Hassan, B.A. (2012) Two new Kirschsteiniothelia species with Dendryphiopsis anamorphs cluster in Kirschsteiniotheliaceae fam. nov. Mycologia 104: 698–714. https://doi.org/10.3852/11-089
- Bugni, T.S. & Ireland, C.M. (2004) Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Natural Product Reports 21: 143–163. https://doi.org/10.1039/b301926h
- Capella-Gutierrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics (Oxford, England) 25: 1972–3. https://doi.org/10.1093/bioinformatics/btp348
- Chaiwan, N., Gomdola, D., Wang, S., Monkai, J., Tibpromma, S., Doilom, M., Wanasinghe, D.N., Mortimer, P.E., Lumyong, S. & Hyde, K.D. (2021) https://gmsmicrofungi.org: an online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere 12: 1513–1526. https://doi.org/10.5943/mycosphere/12/1/19
- Chethana, K.W.T., Manawasinghe, I.S., Hurdeal, V.G., Bhunjun, C.S., Appadoo, M.A., Gentekaki, E., Raspé, O., Promputtha, I. & Hyde, K.D. (2021) What are fungal species and how to delineate them? Fungal Diversity 109: 1–25. https://doi.org/10.1007/s13225-021-00483-9
- Darriba, D., Posada, D., Kozlov, A.M., Stamatakis, A., Morel, B. & Flouri, T. (2020) ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Molecular Biology and Evolution 37: 291–294. https://doi.org/10.1093/molbev/msz189
- Guegan, H., Cailleaux, M., Le Gall, F., Robert-Gangneux, F. & Gangneux, J.P. (2021) Chromoblastomycosis due to a never-before-seen dematiaceous fungus in a kidney transplant patient. Microorganisms 9: 2139. https://doi.org/10.3390/microorganisms9102139
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. [London]: Information Retrieval Ltd., c1979-c2000., pp. 95–98.
- Hawksworth, D.L. (1985) Kirschsteiniothelia, a new genus for the Microthelia incrustans-group (Dothideales). Botanical Journal of the Linnean Society 91: 181–202. https://doi.org/10.1111/j.1095-8339.1985.tb01144.x
- Hernández-Restrepo, M., Gené, J., Castañeda-Ruiz, R.F., Mena-Portales, J., Crous, P.W. & Guarro, J. (2017) Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. https://doi.org/10.1016/j.simyco.2017.05.002
- Hoang, D.T., Chernomor, O., Von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2017) UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281
- Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H.C., Sarma, V.V., Boonmee, S., Lücking, R., Bhat, D.J., Liu, N.G., Tennakoon, D.S., Pem, D., Karunarathna, A., Jiang, S.H., Jones, E.B.G., Phillips, A.J.L., Manawasinghe, I.S., Tibpromma, S., Jayasiri, S.C., Sandamali, D.S., Jayawardena, R.S., Wijayawardene, N.N., Ekanayaka, A.H., Jeewon, R., Lu, Y.Z., Dissanayake, A.J., Zeng, X.Y., Luo, Z.L., Tian, Q., Phukhamsakda, C., Thambugala, K.M., Dai, D.Q., Chethana, K.W.T., Samarakoon, M.C., Ertz, D., Bao, D.F., Doilom, M., Liu, J.K., Pérez-Ortega, S., Suija, A., Senwanna, C., Wijesinghe, S.N., Konta, S., Niranjan, M., Zhang, S.N., Ariyawansa, H.A., Jiang, H.B., Zhang, J.F., Norphanphoun, C., Silva, N.I. de, Thiyagaraja, V., Zhang, H., Bezerra, J.D.P., Miranda-González, R., Aptroot, A., Kashiwadani, H., Harishchandra, D., Sérusiaux, E., Aluthmuhandiram, J.V.S., Abeywickrama, P.D., Devadatha, B., Wu, H.X., Moon, K.H., Gueidan, C., Schumm, F., Bundhun, D., Mapook, A., Monkai, J., Chomnunti, P., Suetrong, S., Chaiwan, N., Dayarathne, M.C., Yang, J., Rathnayaka, A.R., Bhunjun, C.S., Xu, J.C., Zheng, J.S., Liu, G., Feng, Y. & Xie, N. (2020) Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11: 1553–2107. https://doi.org/10.5943/mycosphere/11/1/13
- Hyde, K.D., Chaiwan, N., Norphanphoun, C., Boonmee, S., Erio, C., Chethana, K.W.T., Dayarathne, M.C., de Silva, N.I., Dissanayake, A.J., Ekanayaka, A.H., Hongsanan, S., Huang, S.K., Jayasiri, S.C., Jayawardena, R.S., Jiang, H.B., Karunarathna, A., Lin, C.G., Liu, J.K., Liu, N.G., Lu, Y.Z., Luo, Z.L., Maharachchikumbura, S.S.N., Manawasinghe, I.S., Pem, D., Perera, R.H., Phukhamsakda, C., Samarakoon, M.C., Senwanna, C., Shang, Q.J., Tennakoon, D.S., Thambugala, K.M., Tibpromma, S., Wanasinghe, D.N., Xiao, Y.P., Yang, J., Zeng, X.Y., Zhang, J.F., Zhang, S.N., Bulgakov, T.S., Bhat, D.J., Cheewangkoon, R., Goh, T.K., Jones, E.B.G., Kang, J.C., Jeewon, R., Liu, Z.Y., Lumyong, S., Kuo, C.H., McKenzie, E.H.C., Wen, T.C., Yan, J.Y. & Zhao, Q. (2018) Mycosphere notes 169–224. Mycosphere 9: 271–430. https://doi.org/10.5943/mycosphere/9/2/8
- Hyde, K.D., Jones, E.B.G., Liu, J.K., Ariyawansa, H., Boehm, E., Boonmee, S., Braun, U., Chomnunti, P., Crous, P.W., Dai, D.Q., Diederich, P., Dissanayake, A., Doilom, M., Doveri, F., Hongsanan, S., Jayawardena, R., Lawrey, J.D., Li, Y.M., Liu, Y.X., Lücking, R., Monkai, J., Muggia, L., Nelsen, M.P., Pang, K.L., Phookamsak, R., Senanayake, I.C., Shearer, C.A., Suetrong, S., Tanaka, K., Thambugala, K.M., Wijayawardene, N.N., Wikee, S., Wu, H.X., Zhang, Y., Aguirre-Hudson, B., Alias, S.A., Aptroot, A., Bahkali, A.H., Bezerra, J.L., Bhat, D.J., Camporesi, E., Chukeatirote, E., Gueidan, C., Hawksworth, D.L., Hirayama, K., De Hoog, S., Kang, J.C., Knudsen, K., Li, W.J., Li, X.H., Liu, Z.Y., Mapook, A., McKenzie, E.H.C., Miller, A.N., Mortimer, P.E., Phillips, A.J.L., Raja, H.A., Scheuer, C., Schumm, F., Taylor, J.E., Tian, Q., Tibpromma, S., Wanasinghe, D.N., Wang, Y., Xu, J.C., Yacharoen, S., Yan, J.Y. & Zhang, M. (2013) Families of Dothideomycetes. Fungal Diversity 63: 1–313. https://doi.org/10.1007/s13225-013-0263-4
- Hyde, K.D., Norphanphoun, C., Abreu, V.P., Bazzicalupo, A., Thilini Chethana, K.W., Clericuzio, M., Dayarathne, M.C., Dissanayake, A.J., Ekanayaka, A.H., He, M.Q., Hongsanan, S., Huang, S.Ke., Jayasiri, S.C., Jayawardena, R.S., Karunarathna, A., Konta, S., Kušan, I., Lee, H., Li, J., Lin, C.Gen., Liu, N.Guo., Lu, Y.Z., Luo, Z.L., Manawasinghe, I.S., Mapook, A., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Siedlecki, I., Soares, A.M., Tennakoon, D.S., Tian, Q., Tibpromma, S., Wanasinghe, D.N., Xiao, Y.P., Yang, J., Zeng, X.Y., Abdel-Aziz, F.A., Li, W.J., Senanayake, I.C., Shang, Q.J., Daranagama, D.A., de Silva, N.I., Thambugala, K.M., Abdel-Wahab, M.A., Bahkali, A.H., Berbee, M.L., Boonmee, S., Bhat, D.J., Bulgakov, T.S., Buyck, B., Camporesi, E., Castañeda-Ruiz, R.F., Chomnunti, P., Doilom, M., Dovana, F., Gibertoni, T.B., Jadan, M., Jeewon, R., Jones, E.B.G., Kang, J.C., Karunarathna, S.C., Lim, Y.W., Liu, J.K., Liu, Z.Y., Plautz, H.L., Lumyong, S., Maharachchikumbura, S.S.N., Matočec, N., McKenzie, E.H.C., Mešić, A., Miller, D., Pawłowska, J., Pereira, O.L., Promputtha, I., Romero, A.I., Ryvarden, L., Su, H.Y., Suetrong, S., Tkalčec, Z., Vizzini, A., Wen, T.C., Wisitrassameewong, K., Wrzosek, M., Xu, J.C., Zhao, Q., Zhao, R.L. & Mortimer, P.E. (2017) Fungal diversity notes 603–708: taxonomic and phylogenetic notes on genera and species. Fungal Diversity 87: 1–235. https://doi.org/10.1007/s13225-017-0391-3
- Hyde, K.D., Norphanphoun, C., Ma, J., Yang, H.D., Zhang, J.Y., Du, T.Y., Gao, Y., Gomes De Farias, A.R., Gui, H., He, S.C., He, Y.K., Li, C.J.Y., Liu, X.F., Lu, L., Su, H.L., Tang, X., Tian, X.G., Wang, S.Y., Wei, D.P., Xu, R.F., Xu, R.J., Yang, Y.Y., Zhang, F., Zhang, Q., Bahkali, A.H., Boonmee, S., Chethana, K.W.T., Jayawardena, R.S., Lu, Y.Z., Karunarathna, S.C., Tibpromma, S., Wang, Y. & Zhao, Q. (2023) Mycosphere notes 387–412 – novel species of fungal taxa from around the world. Mycosphere 14: 663–744. https://doi.org/10.5943/mycosphere/14/1/8
- Index Fungorum (2023) Available from: http://indexfungorum.org/ (Accessed: 19 September 2023).
- Jayasiri, S.C., Hyde, K.D., Ariyawansa, H.A., Bhat, J., Buyck, B., Cai, L., Dai, Y.-C., Abd-Elsalam, K.A., Ertz, D. & Hidayat, I. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal diversity 74: 3–18. https://doi.org/10.1007/s13225-015-0351-8
- Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualisation. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
- Larget, B. & Simon, D.L. (1999) Markov Chain Monte Carlo Algorithms for the Bayesian Analysis of Phylogenetic Trees. Molecular Biology and Evolution 16: 750–759. https://doi.org/10.1093/oxfordjournals.molbev.a026160
- Li, G.J., Hyde, K.D., Zhao, R.L., Hongsanan, S., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Alvarado, P., Alves-Silva, G., Ammirati, J.F., Ariyawansa, H.A., Baghela, A., Bahkali, A.H., Beug, M., Bhat, D.J., Bojantchev, D., Boonpratuang, T., Bulgakov, T.S., Camporesi, E., Boro, M.C., Ceska, O., Chakraborty, D., Chen, J.J., Chethana, K.W.T., Chomnunti, P., Consiglio, G., Cui, B.K., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Das, K., Dayarathne, M.C., De Crop, E., De Oliveira, R.J.V., de Souza, C.A.F., de Souza, J.I., Dentinger, B.T.M., Dissanayake, A.J., Doilom, M., Drechsler-Santos, E.R., Ghobad-Nejhad, M., Gilmore, S.P., Góes-Neto, A., Gorczak, M., Haitjema, C.H., Hapuarachchi, K.K., Hashimoto, A., He, M.Q., Henske, J.K., Hirayama, K., Iribarren, M.J., Jayasiri, S.C., Jayawardena, R.S., Jeon, S.J., Jerônimo, G.H., Jesus, A.L., Jones, E.B.G., Kang, J.C., Karunarathna, S.C., Kirk, P.M., Konta, S., Kuhnert, E., Langer, E., Lee, H.S., Lee, H.B., Li, W.J., Li, X.H., Liimatainen, K., Lima, D.X., Lin, C.G., Liu, J.K., Liu, X.Z., Liu, Z.Y., Luangsa-ard, J.J., Lücking, R., Lumbsch, H.T., Lumyong, S., Leaño, E.M., Marano, A.V., Matsumura, M., McKenzie, E.H.C., Mongkolsamrit, S., Mortimer, P.E., Nguyen, T.T.T., Niskanen, T., Norphanphoun, C., O’Malley, M.A., Parnmen, S., Pawłowska, J., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Pires-Zottarelli, C.L.A., Raspé, O., Reck, M.A., Rocha, S.C.O., de Santiago, A.L.C.M.A., Senanayake, I.C., Setti, L., Shang, Q.J., Singh, S.K., Sir, E.B., Solomon, K.V., Song, J., Srikitikulchai, P., Stadler, M., Suetrong, S., Takahashi, H., Takahashi, T., Tanaka, K., Tang, L.P., Thambugala, K.M., Thanakitpipattana, D., Theodorou, M.K., Thongbai, B., Thummarukcharoen, T., Tian, Q., Tibpromma, S., Verbeken, A., Vizzini, A., Vlasák, J., Voigt, K., Wanasinghe, D.N., Wang, Y., Weerakoon, G., Wen, H.A., Wen, T.C., Wijayawardene, N.N., Wongkanoun, S., Wrzosek, M., Xiao, Y.P., Xu, J.C., Yan, J.Y., Yang, J., Da Yang, S., Hu, Y., Zhang, J.F., Zhao, J., Zhou, L.W., Peršoh, D., Phillips, A.J.L. & Maharachchikumbura, S.S.N. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. https://doi.org/10.1007/s13225-016-0366-9
- Maharachchikumbura, S.S.N., Chen, Y., Ariyawansa, H.A., Hyde, K.D., Haelewaters, D., Perera, R.H., Samarakoon, M.C., Wanasinghe, D.N., Bustamante, D.E., Liu, J.K., Lawrence, D.P., Cheewangkoon, R. & Stadler, M. (2021) Integrative approaches for species delimitation in Ascomycota. Fungal Diversity 109: 155–179. https://doi.org/10.1007/s13225-021-00486-6
- Mehrabi, M., Hemmati, R. & Asgari, B. (2017) Kirschsteiniothelia arasbaranica sp. nov., and an Emendation of the Kirschsteiniotheliaceae. Cryptogamie Mycologie 38: 13–25. https://doi.org/10.7872/crym/v38.iss1.2017.13
- Miller, M., Schwartz, T., Pickett, B., He, S., Klem, E., Scheuermann, R., Passarotti, M., Kaufman, S. & O’Leary, M. (2015) A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evolutionary bioinformatics online 2015: 43–48. https://doi.org/10.4137/EBO.S21501
- Nguyen, L.T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. https://doi.org/10.1093/molbev/msu300
- Nishi, M., Okano, I., Sawada, T., Hara, Y., Nakamura, K., Inagaki, K. & Yaguchi, T. (2018) Chronic Kirschsteiniothelia infection superimposed on a pre-existing non-infectious bursitis of the ankle: the first case report of human infection. BMC Infectious Diseases 18: 236. https://doi.org/10.1186/s12879-018-3152-3
- Poch, G.K., Gloer, J.B. & Shearer, C.A. (1992) New bioactive metabolites from a freshwater isolate of the fungus Kirschsteiniothelia sp. Journal of Natural Products 55: 1093–1099. https://doi.org/10.1021/np50086a010
- Rambaut, A. (2012) FigTree version 1.4.4. Available from: http://tree.bio.ed.ac.uk/software/figtree/ (Accessed 15 May 2023)
- Rehner, S.A. & Samuels, G.J. (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625–634. https://doi.org/10.1016/S0953-7562(09)80409-7
- Sayers, E.W., Cavanaugh, M., Clark, K., Ostell, J., Pruitt, K.D. & Karsch-Mizrachi, I. (2020) GenBank. Nucleic Acids Research 48: D84–D86. https://doi.org/10.1093/nar/gkz956
- Schoch, C.L., Crous, P.W., Groenewald, J.Z., Boehm, E.W.A., Burgess, T.I., de Gruyter, J., de Hoog, G.S., Dixon, L.J., Grube, M., Gueidan, C., Harada, Y., Hatakeyama, S., Hirayama, K., Hosoya, T., Huhndorf, S.M., Hyde, K.D., Jones, E.B.G., Kohlmeyer, J., Kruys, Å., Li, Y.M., Lücking, R., Lumbsch, H.T., Marvanová, L., Mbatchou, J.S., McVay, A.H., Miller, A.N., Mugambi, G.K., Muggia, L., Nelsen, M.P., Nelson, P., Owensby, C.A., Phillips, A.J.L., Phongpaichit, S., Pointing, S.B., Pujade-Renaud, V., Raja, H.A., Plata, E.R., Robbertse, B., Ruibal, C., Sakayaroj, J., Sano, T., Selbmann, L., Shearer, C.A., Shirouzu, T., Slippers, B., Suetrong, S., Tanaka, K., Volkmann-Kohlmeyer, B., Wingfield, M.J., Wood, A.R., Woudenberg, J.H.C., Yonezawa, H., Zhang, Y. & Spatafora, J.W. (2009) A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. https://doi.org/10.3114/sim.2009.64.01
- Senanayake, I., Rathnayaka, A., Sandamali, D., Calabon, M., Gentekaki, E., Lee, H., Pem, D., Dissanayake, L., Wijesinghe, S., Bundhun, D., TTT, N., Goonasekara, I., Abeywickrama, P., Jayawardena, R., Wanasinghe, D., Jeewon, R., Bhat, D.J., Mm, X., Bhunjun, C.S. & Hurdeal, V. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
- Su, H., Hyde, K.D., Maharachchikumbura, S.S.N., Ariyawansa, H.A., Luo, Z., Promputtha, I., Tian, Q., Lin, C., Shang, Q., Zhao, Y., Chai, H., Liu, X., Bahkali, A.H., Bhat, J.D., McKenzie, E.H.C. & Zhou, D. (2016) The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Diversity 80: 375–409. https://doi.org/10.1007/s13225-016-0362-0
- Sun, Y.-R., Jayawardena, R.S., Hyde, K.D. & Wang, Y. (2021) Kirschsteiniothelia thailandica sp. nov. (Kirschsteiniotheliaceae) from Thailand. Phytotaxa 490: 172–182. https://doi.org/10.11646/phytotaxa.490.2.3
- Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
- White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322.
- Wijayawardene, N.N., Crous, P.W., Kirk, P.M., Hawksworth, D.L., Boonmee, S., Braun, U., Dai, D.-Q., D’souza, M.J., Diederich, P., Dissanayake, A., Doilom, M., Hongsanan, S., Jones, E.B.G., Groenewald, J.Z., Jayawardena, R., Lawrey, J.D., Liu, J.K., Lücking, R., Madrid, H., Manamgoda, D.S., Muggia, L., Nelsen, M.P., Phookamsak, R., Suetrong, S., Tanaka, K., Thambugala, K.M., Wanasinghe, D.N., Wikee, S., Zhang, Y., Aptroot, A., Ariyawansa, H.A., Bahkali, A.H., Bhat, D.J., Gueidan, C., Chomnunti, P., De Hoog, G.S., Knudsen, K., Li, W.J., McKenzie, E.H.C., Miller, A.N., Phillips, A.J.L., Piątek, M., Raja, H.A., Shivas, R.S., Slippers, B., Taylor, J.E., Tian, Q., Wang, Y., Woudenberg, J.H.C., Cai, L., Jaklitsch, W.M. & Hyde, K.D. (2014) Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Diversity 69: 1–55. https://doi.org/10.1007/s13225-014-0309-2
- Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sánchez-García, M., Goto, B.T., Saxena, R.K., Erdoðdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., Błaszkowski, J., Boonyuen, N., da Silva, G.A., de Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Mešić, A., Monteiro, J.S., Oehl, F., Pawłowska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkalčec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro, I., Tang, L., Stadler, M., Yurkov, A. & Thines, M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13: 53–453. https://doi.org/10.5943/mycosphere/13/1/2
- Wu, K., Liu, Y., Liao, X., Yang, X., Chen, Z., Mo, L., Zhong, S. & Zhang, X. (2023) Fungal Diversity and Its Relationship with Environmental Factors in Coastal Sediments from Guangdong, China. Journal of Fungi 9: 101. https://doi.org/10.3390/jof9010101
- Xu, R., Phukhamsakda, C., Dai, D., Karunarathna, S. & Tibpromma, S. (2023) Kirschsteiniothelia xishuangbannaensis sp. nov. from pará rubber (Hevea brasiliensis) in Yunnan, China. Current Research in Environmental & Applied Mycology 13: 34–56. https://doi.org/10.5943/cream/13/1/3
- Yang, J., Liu, L.-L., Jones, E.B.G., Hyde, K.D., Liu, Z.-Y., Bao, D.-F., Liu, N.-G., Li, W.-L., Shen, H.-W., Yu, X.-D. & Liu, J.-K. (2023) Freshwater fungi from karst landscapes in China and Thailand. Fungal Diversity 119: 1–212. https://doi.org/10.1007/s13225-023-00514-7