Abstract
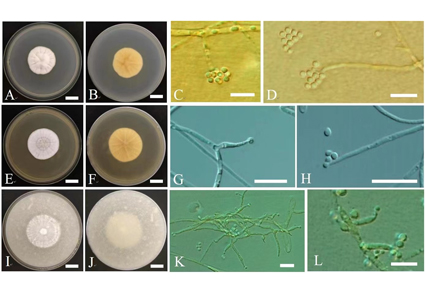
Tolypocladium is a diverse group of asexual or sexual fungi within the Ophiocordycipitaceae family, exhibiting a diverse range of hosts, living habits and geographical distribution. In this study, a Tolypocladium strain RCEF6720 was isolated from the rhizome of Polygonatum cyrtonema, collected in Huangshan City, Anhui Province, China. Phylogenetic analyses of multiple loci (ITS, SSU, LSU, TEF1-α and RPB1) indicated that the strain RCEF6720 was clustered with other Tolypocladium species associated with plant habitats, including T. album, T. amazonense, T. endophyticum and T. tropicale. Further study revealed that the strain RCEF6720 was identified as a distinct lineage, separated from T. album. While the strain’s morphological characteristics resemble those of T. album, it is distinguished by longer phialides. Based on the combination of its unique morphological characteristics and phylogenetic placement, the strain is proposed as a new species, named Tolypocladium rhizomatum sp. nov.
References
- Bischoff, J.F.& White, J.F. (2004) Torrubiella piperis sp. nov. (Clavicipitaceae, Hypocreales), a new teleomorph of the Lecanicillium complex. Studies in Mycology 50 (1): 89–94. https://doi.org/10.1023/B:MYCO.0000012225.79969.29
- Bischoff, J.F., Rehner, S.A. & Humber, R.A. (2009) A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 101: 512–530. https://doi.org/10.3852/07-202
- Chen, X.J., Duan, J.F., Liu, K.Q., Guo, Y.Y., Wang, D.P., Liu, M., Zhao, D., Li, B., Li, H.L. & Wang, X.B. (2021) Botany, traditional uses, and pharmacology of Polygonati rhizoma. Chinese Medicine and Culture 4 (4): 251–259. https://doi.org/10.4103/cmac.cmac_39_21
- Chernomor, O., Haeseler, A.V. & Minh, B.Q. (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65 (6): 997–1008. https://doi.org/10.1093/sysbio/syw037
- Faeth, S.H. & Fagan, W.F. (2002) Fungal endophytes: common host plant symbionts but uncommon mutualists. Integrative and Comparative Biology 42 (2): 360–368. https://doi.org/10.1093/icb/42.2.360
- Fukuda, T., Sudoh, Y., Tsuchiya, Y., Okuda, T., Matsuura, N., Motojima, A., Oikawa, T. & Lgarashi, Y. (2015) Tolypoalbin, a new tetramic acid from Tolypocladium album TAMA 479. The Journal of Antibiotics 68: 399–402. https://doi.org/10.1038/ja.2014.165
- Gallardo-Pillancari, E., Gonzalez, C., Barahona-Segovia, R.M., Ruiz, C., Luz, C., Humber, R.A. & Montalva, C. (2023) Natural infection of Chiromyzinae larvae (Diptera: Stratiomyidae) in southern Chile by Tolypocladium valdiviae sp. nov. Fungal Biology 127: 845–853. https://doi.org/10.1016/j.funbio.2022.12.004
- Gams, W. (1971) Tolypocladium, eine Hyphomycetengattung mit geschwollenen Phialiden. Persoonia-Molecular Phylogeny and Evolution of Fungi 6 (2): 185–191.
- Gams, W. (1980) Chaunopycnis alba, gen. et sp. nov., a soil fungus intermediate between Moniliales and Sphaeropsidales. Persoonia - Molecular Phypgeny and Evolution of Fungi 11: 75–79.
- Gazis, R., Skaltsas, D. & Chaverri, P. (2014) Novel endophytic lineages of Tolypocladium provide new insights into the ecology and evolution of Cordyceps-like fungi. Mycologia 106 (6): 1090–1105. https://doi.org/10.3852/13-346
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology 59 (3): 307–321. https://doi.org/10.1093/sysbio/syq010
- Hall, T., Biosciences, I. & Carlsbad, C. (2011) BioEdit: an important software for molecular biology. GERF Bulletin of Biosciences 2 (1): 60–61. [https://www.researchgate.net/publication/258565830]
- Hoang, D.T., Chernomor, O., Arndt, V.H., Quang, M.B. & Sy, V.L. (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35 (2): 518–22. https://doi.org/10.1093/molbev/msx281
- Jie, A.N., Bao, J.K., Liu, J.Z., Chuan-Fang, W.U., Jian, L.I., Lei, D., Balzarini, J., Clercq, E.D. & Fang, C. (2006) Anti‐HIV I/II activity and molecular cloning of a novel mannose/sialic acid‐binding lectin from rhizome of Polygonatum cyrtonema Hua. Acta Biochimica et Biophysica Sinica 38 (2): 70–78. https://doi.org/10.1111/j.1745-7270.2006.00140
- Kaul, S., Gupta, S., Ahmed, M. & Manoj, K.D. (2012) Endophytic fungi from medicinal plants: a treasure hunt for bioactive metabolites. Phytochemistry Reviews 11 (4): 487–505. https://doi.org/10.1007/s11101-012-9260-6
- Kneifel, H., Konig, W.A., Loeffler, W. & Muller, R. (1977) Ophiocordin, an antifungal antibiotic of Cordyceps ophioglossoides. Archives of Microbiology 113: 121–130. https://doi.org/10.1007/BF00428591
- Kranoff, S.B. & Gupta, S. (1992) Efrapeptin production by Tolypocladium fungi (Deuteromycotina: Hyphomycetes): Intra-and interspecific variation. Journal of Chemical Ecology 18 (10): 1727–1741. https://doi.org/10.1007/BF02751098
- Liu, Y.Z (2015) Polygonatum cyrtonema Hua. In: Liu, Y.Z, Wang, Z.M. & Zhang, J.Z. (eds.) Dietary Chinese herbs. Springer, Vienna, pp. 213–218. https://doi.org/10.1007/978-3-211-99448-1
- Liu, Z.Y., Liang, Z.Q., Whalley, A.J.S., Yao, Y.J. & Liu, A.Y. (2001) Cordyceps brittlebankisoides, a new pathogen of grubs and its anamorph, Metarhizium anisopliae var. majus. Journal of Invertebrate Pathology 78 (3): 178–182.
- https://doi.org/10.1006/jipa.2001.5039
- Namasivayam, K.R., Swetha, R. & Srivatsan, K.V. (2014) Evaluation of potential biological activities of metabolites from endophytic fungi residing in leaves of Azadirhacta indica. International Journal of ChemTech Research 6 (5): 3116–3121. [https://www.researchgate.net/publication/265729353]
- Nguyen, L.T., Schmidt, H.A., Haeseler, A.V. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32 (1): 268–274.
- https://doi.org/10.1093/molbev/msu300
- Quandt, C.A., Kepler, R.M., Gams, W., Araujo, J.P.M., Ban, S., Evans, H., Hughes, D., Humber, R., Hywel-Jones, N., Li, Z.Z., Luangsa-ard, J.J., Rehner, S.A., Sanjuan, T., Sato, H., Shrestha, B., Sung, G.H., Yao, Y.J., Zare, R. & Spatafora, J.W. (2014) Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus 5 (1): 121–134. https://doi.org/10.5598/imafungus.2014.05.01.12
- Rehner, S.A. & Buckley, E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97 (1): 84–98.
- https://doi.org/10.3852/mycologia.97.1.84
- Rehner, S.A., Minnis, A.M., Sung, G.H., Luangsa-ard, J.J., Devotto, L. & Humber, R.A. (2011) Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103 (5): 1055–1073. https://doi.org/10.3852/10-302
- Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 (12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
- Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–542. https://doi.org/10.1093/sysbio/sys029
- Schulz, B., Boyle, C., Draeger, S., Rommert, A.K. & Krohn, K. (2002) Endophytic fungi: a source of novel biologically active secondary metabolites. Mycological Research 106 (9): 996–1004. https://doi.org/10.1017/S0953756202006342
- Smithe, F.B. (1975) Naturalist’s color guide[M]. American Museum of Natural History (New York).
- Wang, Y.B., Yu, H., Dai, Y.D., Wu, C.K., Zeng, W.B., Yuan, F. & Liang, Z.Q. (2015) Polycephalomyces agaricus, a new hyperparasite of Ophiocordyceps sp. infecting melolonthid larvae in southwestern China. Mycological Progress 17: 745–753. https://doi.org/10.1007/s11557-017-1370-5
- Wang, J.C., Zhang, Z.Z., Li, Z.L. & Wang, Y. (2020) Research progress of Tolypocladium in Ophiocordycipitaceae. Journal of Fungal Research 18 (01): 54–62. https://doi.org/10.13341/j.jfr.2020.1259
- Wang, Y.B., Wang, Y., Fan, Q., et al. (2020) Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepialid. Fungal diversity 103: 1–46. https://doi.org/10.1007/s13225-020-00457-3
- White, T.J., Bruns, T.D., Lee, S.B. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18 (1): 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wijayawardene, N.N., Hyde, K.D., Rajeshkumar, K.C., Hawksworth, D.L., Madrid, H., Kirk, P.M., Braun, U., Singh, R.V., Crous, P.W., Kukwa, M., Lücking, R., Kurtzman, C.P., Yurkov, A., Haelewaters, D., Aptroot, A., Lumbsch, H.T., Timdal, E., Ertz, D., Etayo, J., Phillips, A.J.L., Groenewald, J.Z., Papizadeh, M., Selbmann, L., Dayarathne, M.C., Weerakoon, G., Jones, E.B.G., Suetrong, S., Tian, Q., Castañeda-Ruiz, R.F., Bahkali, A.H., Pang, K.-L., Tanaka, K., Dai, D.Q., Sakayaroj, J., Hujslová, M., Lombard, L., Shenoy, B.D., Suija, A., Maharachchikumbura, S.S.N., Thambugala, K.M., Wanasinghe, D.N., Sharma, B.O., Gaikwad, S., Pandit, G., Zucconi, L., Onofri, S., Egidi, E., Raja, H.A., Kodsueb, R., Cáceres, M.E.S., Pérez-Ortega, S., Fiuza, P.O., Monteiro, J.S., Vasilyeva, L.N., Shivas, R.G., Prieto, M., Wedin, M., Olariaga, I., Lateef, A.A., Agrawal, Y., Fazeli, S.A.S., Amoozegar, M.A., Zhao, G.Z., Pfliegler, W.P., Sharma, G., Oset, M., Abdel-Wahab, M.A., Takamatsu, S., Bensch, K., de Silva, N.I., De Kesel, A., Karunarathna, A., Boonmee, S., Pfister, D.H., Lu, Y.-Z., Luo, Z.-L., Boonyuen, N., Daranagama, D.A., Senanayake, I.C., Jayasiri, S.C., Samarakoon, M.C., Zeng, X.-Y., Doilom, M., Quijada, L., Rampadarath, S., Heredia, G., Dissanayake, A.J., Jayawardana, R.S., Perera, R.H., Tang, L.Z., Phukhamsakda, C., Hernández-Restrepo, M., Ma, X., Tibpromma, S., Gusmao, L.F.P., Weerahewa, D. & Karunarathna, S.C. (2017) Notes for genera: Ascomycota. Fungal diversity 86 (1): 1–594. https://doi.org/10.1007/s13225-017-0386-0
- Wijayawardene, N.N., Hyde, K.D., Al-Ani, L. & Tedersoo, L. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11 (1): 1160–1456. https://doi.org/10.5943/mycosphere/11/1/8
- Wijayawardene, N.N., Dissanayake, L.S., Li, Q.-R., Dai, D.-Q., Xiao, Y., Wen, T.-C., Karunarathna, S.C., Wu, H.-X., Zhang, H., Tibpromma, S., Kang, J.-C., Wang, Y., Shen, X.-C., Tang, L.-Z., Deng, C.-Y., Liu, Y. & Kang, Y. (2021) Yunnan-Guizhou Plateau: a mycological hotspot. Phytotaxa 523 (1): 1–31. https://doi.org/10.11646/phytotaxa.523.1.1
- Yamamoto, K., Sugawa, G., Takeda, K., Degawa, Y. (2022) Tolypocladium bacillisporum (Ophiocordycepitaceae): A new parasite of Elaphomyces from Japan. Truffology 5 (1): 15–21. [https://jats-truffles.org/truffology/]
- Yu, F.M., Thilini Chethana, K.W., Wei, D.P., Liu, J.W., Zhao, Q., Tang, S.M., Li, L. & Hyde, K.D. (2021) Comprehensive review of Tolypocladium and description of a novel lineage from southwest China. Pathogens 10 (11): 1389–1402. https://doi.org/10.3390/pathogens10111389