Abstract
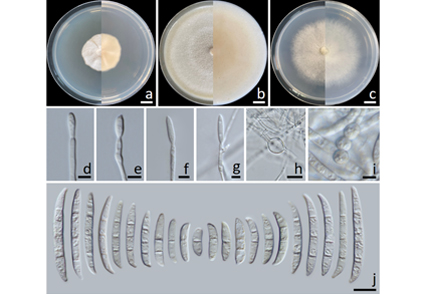
A new endophytic ascomycete, described herein as Fusarium endophyticum, was isolated from leaves of Camellia sinensis var. assamica collected from Nan Province, northern Thailand. This species is described using morphological characteristics and molecular multi-gene analyses. Multi-gene phylogenetic analyses of a combination of internal transcribed spacer region of the rDNA (ITS), translation-elongation factor 1 alpha (tef-1), RNA polymerase II second largest subunit (rpb2), and RNA polymerase II largest subunit (rpb1) genes revealed that F. endophyticum belonged to the Fusarium lateritium species complex (FLSC) and was distinct from all other known species. A full description, illustrations and a phylogenetic tree showing the position of F. endophyticum are provided.
References
- Abi Saad, C., Masiello, M., Habib, W., Gerges, E., Sanzani, S.M., Logrieco, A.F., Moretti, A. & Somma, S. (2022) Diversity of Fusarium species isolated from symptomatic plants belonging to a wide range of agri-food and ornamental crops in Lebanon. Journal of Fungi 8: 897. https://doi.org/10.3390/jof8090897
- Ajmal, M., Hussain, A., Ali, A., Chen, H. & Lin, H. (2023) Strategies for controlling the sporulation in Fusarium spp. Journal of Fungi 9: 10. https://doi.org/10.3390/jof9010010
- Askun, T. (2018) Introductory chapter: Fusarium: pathogenicity, infections, diseases, mycotoxins and management. In: Askun, T. (ed.) Fusarium - Plant Diseases, Pathogen Diversity, Genetic Diversity, Resistance and Molecular Markers. IntechOpen, London, United Kingdom. https://doi.org/10.5772/intechopen.76507
- Babadoost, M. (2018) Fusarium: historical and continued importance. In: Askun, T. (ed.) Fusarium - Plant Diseases, Pathogen Diversity, Genetic Diversity, Resistance and Molecular Markers. IntechOpen, London, United Kingdom. https://doi.org/10.5772/intechopen.74147
- Balajee, S.A., Borman, A.M., Brandt, M.E., Cano, J., Cuenca-Estrella, M., Dannaoui, E., Guarro, J., Haase, G., Kibbler, C.C., Meyer, W., O’Donnell, K., Petti, C.A., Rodriguez-Tudela, J.L., Sutton, D., Velegraki, A. & Wickes, B.L. (2009) Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: Where are we and where should we go from here? Journal of Clinical Microbiology 47: 877–884. https://doi.org/10.1128/JCM.01685-08
- Cavalcanti, A.D., da Silva Santos, A.C., de Oliveria Ferro, L., Bezerra, J.D.P., Souza-Motta, C.M. & Magalhaes, O.M.C. (2020) Fusarium massalimae sp. nov. (F. lateritium species complex) occurs endophytically in leaves of Handroanthus chrysotrichus. Mycological Progress 19: 1133–1142. https://doi.org/10.1007/s11557-020-01622-3
- Crous, P.W., Lombard, L., Sandoval-Denis, M., Seifert, K.A., Schroers, H.-J., Chaverri, P., Gené, J., Guarro, J., Hirooka, Y., Bensch, K., Kema, G.H.J., Lamprecht, S.C., Cai, L., Rossman, A.Y., Stadler, M., Summerbell, R.C., Taylor, J.W., Ploch, S., Visagie, C.M., Yilmaz, N., Frisvad, J.C., Abdel-Azeem, A.M., Abdollahzadeh, J., Abdolrasouli, A., Akulov, A., Alberts, J.F., Araújo, J.P.M., Ariyawansa, H.A., Bakhshi, M., Bendiksby, M., Ben Hadj Amor, A., Bezerra, J.D.P., Boekhout, T., Câmara, M.P.S., Carbia, M., Cardinali, G., Castañeda-Ruiz, R.F., Celis, A., Chaturvedi, V., Collemare, J., Croll, D., Damm, U., Decock, C.A., de Vries, R.P., Ezekiel, C.N., Fan, X.L., Fernández, N.B., Gaya, E., González, C.D., Gramaje, D., Groenewald, J.Z., Grube, M., Guevara-Suarez, M., Gupta, V.K., Guarnaccia, V., Haddaji, A., Hagen, F., Haelewaters, D., Hansen, K., Hashimoto, A., Hernández-Restrepo, M., Houbraken, J., Hubka, V., Hyde, K.D., Iturriaga, T., Jeewon, R., Johnston, P.R.. Jurjević, Ž., Karalti, Ý., Korsten, L., Kuramae, E.E., Kušan, I., Labuda, R., Lawrence, D.P., Lee, H.B., Lechat, C., Li, H.Y., Litovka, Y.A., Maharachchikumbura, S.S.N., Marin-Felix, Y., Matio Kemkuignou, B., Matočec, N., McTaggart, A.R., Mlčoch, P., Mugnai, L., Nakashima, C., Nilsson, R.H., Noumeur, S.R., Pavlov, I.N., Peralta, M.P., Phillips, A.J.L., Pitt, J.I., Polizzi, G., Quaedvlieg, W., Rajeshkumar, K.C., Restrepo, S., Rhaiem, A., Robert, J., Robert, V., Rodrigues, A.M., Salgado-Salazar, C., Samson, R.A., Santos, A.C.S., Shivas, R.G., Souza-Motta, C.M., Sun, G.Y., Swart, W.J., Szoke, S., Tan, Y.P., Taylor, J.E., Taylor, P.W.J., Tiago, P.V., Váczy, K.Z., van de Wiele, N., van der Merwe, N.A., Verkley, G.J.M., Vieira, W.A.S., Vizzini, A., Weir, B.S., Wijayawardene, N.N., Xia, J.W., Yáñez-Morales, M.J., Yurkov, A., Zamora, J.C., Zare, R., Zhang, C.L. & Thines, M. (2021) Fusarium: More than a node or a foot-shaped basal cell. Studies in Mycology 98: 100116. https://doi.org/10.1016/j.simyco.2021.100116
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. https://doi.org/10.1038/nmeth.2109
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. https://doi.org/10.1093/nar/gkh340
- Ekwomadu, T.I., Akinola, S.A. & Mwanza, M. (2021) Fusarium mycotoxins, their metabolites (free, emerging, and masked), food safety concerns, and health impacts. International Journal of Environmental Research and Public Health 18: 11741. https://doi.org/10.3390/ijerph182211741
- Felsenstein, J. (1985) Confidence intervals on phylogenetics: an approach using bootstrap. Evolution 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
- Geiser, D.M., Aoki, T., Bacon, C.W., Baker, S.E., Bhattacharyya, M.K., Brandt, M.E., Brown, D.W., Burgess, L.W., Chulze, S., Coleman, J.J., Correll, J.C., Covert, S.F., Crous, P.W., Cuomo, C.A., De Hoog, G.S., Pietro, A.D., Elmer, W.H., Epstein, L., Frandsen, R.J.N., Freeman, S., Gagkaeva, T., Glenn, A.E., Gordon, T.R., Gregory, N.F., Hammond-Kosack, K.E., Hanson, L.E., Jímenez-Gasco, M.M., Kang, S., Kistler, H.C., Kuldau, G.A., Leslie, J.F., Logrieco, A., Lu, G., Lysøe, E., Ma, L.-J., McCormick, S.P., Migheli, Q., Moretti, A., Munaut, F., O’Donnell, K., Pfenning, L., Ploetz, R.C., Proctor, R.H., Rehner, S.A., Robert, V.A.R.G., Rooney, A.P., bin Salleh, B., Scandiani, M.M., Scauflaire, J., Short, D.P.G., Steenkamp, E., Suga, H., Summerell, B.A., Sutton, D.A., Thrane, U., Trail, F., Van Diepeningen, A., VanEtten, H.D., Viljoen, A., Waalwijk, C., Ward, T.J., Wingfield, M.J., Xu, J.-R., Yang, X.-B., Yli-Mattila, T. & Zhang, N. (2013) One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 103: 400−408. https://doi.org/10.1094/PHYTO-07-12-0150-LE
- Geiser, D.M., Jiménez-Gasco, M.M., Kang, S., Makalowska, I., Veeraraghavan, N., Ward, T.J., Zhang, N., Kuldau, G.A. & O’Donnell, K. (2004) FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. European Journal of Plant Pathology 110: 473–479. https://doi.org/10.1023/B:EJPP.0000032386.75915.a0
- Geiser, D.M., Lewis Ivey, M.L., Hakiza, G., Juba, J.H. & Miller, S.A. (2005) Gibberella xylarioides (anamorph: Fusarium xylarioides), a causative agent of coffee wilt disease in Africa, is a previously unrecognised member of the G. fujikuroi species complex. Mycologia 97: 191–201. https://doi.org/10.1080/15572536.2006.11832853
- Gerlach, W. & Nirenberg, H.I. (1982) The genus Fusarium—a pictorial atlas. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 209: 1–406.
- Gräfenhan, T., Schroers, H.-J., Nirenberg, H.I. & Seifert, K.A. (2011) An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Studies in Mycology 68: 79–113. https://doi.org/10.3114/sim.2011.68.04
- Hall, T. (2004) BioEdit. Ibis Therapeutics, Carlsbad, CA, 92008, USA. Available from: http://www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed December 2021 )
- Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4
- Khuna, S., Kumla, J., Thitla, T., Nuangmek, W., Lumyong, S. & Suwannarach, N. (2022) Morphology, molecular identification, and pathogenicity of two novel Fusarium species associated with postharvest fruit rot of cucurbits in northern Thailand. Journal of Fungi 8: 1135. https://doi.org/10.3390/jof8111135
- Leslie, J.F. & Summerell, B.A. (2006) The Fusarium Laboratory Manual, 1st ed. Blackwell Publishing Professional, Ames, IA, USA. pp. 8–240.
- Link, H.F. (1809) Observationes in ordines plantarum naturales. Dissertatio I. Magazin der Gesellschaft Naturforschenden Freunde Berlin 3: 3–42.
- Nuangmek, W., Kumla, J., Khuna, S., Lumyong, S. & Suwannarach, N. (2023) Identification and characterization of Fusarium species causing watermelon fruit rot in northern Thailand. Plants 12: 956. https://doi.org/10.3390/plants12040956
- O’Donnell, K., Kistler, H.C., Cigelnik, E. & Ploetz, R.C. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95: 2044–2049. https://doi.org/10.1073/pnas.95.5.2044
- O’Donnell, K., Rooney, A.P., Proctor, R.H., Brown, D.W., McCormick, S.P., Ward, T.J., Frandsen, R.J., Lysøe, E., Rehner, S.A., Aoki, T., Robert, V.A.R.G., Crous, P.W., Groenewald, J.Z., Kang, S. & Geiser, D.M. (2013) Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology 52: 20–31. https://doi.org/10.1016/j.fgb.2012.12.004
- O’Donnell, K., Sutton, D.A., Rinaldi, M.G., Sarver, B.A.J., Balajee, S.A., Schroers, H.-J., Summerbell, R.C., Robert, V.A.R.G., Crous, P.W., Zhang, N., Aoki, T., Jung, K., Park, J., Lee, Y.-H., Kang, S., Park, B. & Geiser, D.M. (2010) Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology 48: 3708–3718. https://doi.org/10.1128/JCM.00989-10
- O’Donnell, K., Ward, T.J., Robert, V.A.R.G., Crous, P.W., Geiser, D.M. & Kang, S. (2015) DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 43: 583–595. https://doi.org/10.1007/s12600-015-0484-z
- Perera, R.H., Hyde, K.D., Maharachchikumbura, S.S.N., Jones, E.B.G., McKenzie, E.H.C., Stadler, M., Lee, H.B., Samarakoon, M.C., Ekanayaka, A.H., Camporesi, E., Liu, J.K. & Liu, Z.Y. (2020) Fungi on wild seeds and fruits. Mycosphere 11: 2108–2480. https://doi.org/10.5943/mycosphere/11/1/14
- Rahjoo, V., Zad, J., Javan-Nikkhah, M., Mirzadi Gohari, A., Okhovvat, S.M., Bihamta, M.R., Razzaghian, J. & Klemsdal, S.S. (2008) Morphological and molecular identification of Fusarium isolated from maize ears in Iran. Journal of Plant Pathology 90: 463–468.
- Rambaut, A. (2012) FigTree Tree Figure Drawing Tool Version 131, Institute of Evolutionary 623 Biology, University of Edinburgh, Edinburgh, Scotland. Available from: http://treebioedacuk/software/figtree/ (accessed March 2023).
- Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
- Sandoval-Denis, M., Guarnaccia, V., Polizzi, G. & Crous, P.W. (2018) Symptomatic Citrus trees reveal a new pathogenic lineage in Fusarium and two new Neocosmospora species. Persoonia 40: 1–25. https://doi.org/10.3767/persoonia.2018.40.01
- Schroers, H.-J., Gräfenhan, T., Nirenberg, H.I. & Seifert, K.A. (2011) A revision of Cyanonectria and Geejayessia gen. nov., and related species with Fusarium-like anamorphs. Studies in Mycology 68: 115–138. https://doi.org/10.3114/sim.2011.68.05
- Sharma, L. & Marques, G. (2018) Fusarium, an entomopathogen—A myth or reality? Pathogens 7: 93. https://doi.org/10.3390/pathogens7040093
- Stamatakis, A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. https://doi.org/10.1093/bioinformatics/btl446
- Summerell, B.A. (2019) Resolving Fusarium: current status of the genus. Annual Review of Phytopathology 57: 323–339. https://doi.org/10.1146/annurev-phyto-082718-100204
- Suwannarach, N., Bussaban, B., Nuangmek, W., McKenzie, E.H.C., Hyde, K.D. & Lumyong, S. (2012) Diversity of endophytic fungi associated with Cinnamomum bejolghota (Lauraceae) in northern Thailand. Chiang Mai Journal of Science 39: 389−398.
- Tamura, K., Stecher, G., Petersen, D., Filipsky, A. & Kumar, S. (2013) MEGE 6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolutionary 30: 2725–2729. https://doi.org/10.1093/molbev/mst197
- Tshilenge, L., Kalonji, A. & Tshilenge, P. (2010) Determination of cultural and biometrical characters of Fusarium species isolated from plant material harvested from coffee (Coffea canephora Pierre.) infected with CWD in Democratic Republic of Congo. African Journal of Agricultural Research 5: 3145–3150.
- Vitale, S., Santori, A., Wajnberg, E., Castagnone-Sereno, P., Luongo, L. & Belisario, A. (2011) Morphological and molecular analysis of Fusarium lateritium, the cause of gray necrosis of hazelnut fruit in Italy. Phytopathology 101: 679–686. https://doi.org/10.1094/PHYTO-04-10-0120.
- Wang, M.M., Chen, Q., Diao, Y.Z., Duan, W.J. & Cai, L. (2019) Fusarium incarnatum-equiseti complex from China. Persoonia 43: 70–89. https://doi.org/10.3767/persoonia.2019.43.03
- Wang, M.M., Crous, P.W., Sandoval-Denis, M., Han, S.L., Liu, F., Liang, J.M., Duan, W.J. & Cai, L. (2022) Fusarium and allied genera from China: species diversity and distribution. Persoonia 48: 1–53. https://doi.org/10.3767/persoonia.2022.48.01
- White, T.J., Burns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & Whitish, T.J. (Eds.) PCR Protocols, a Guide to Methods and Applications. Academic Press, San Diego. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
- Wollenweber, H.W. (1917) Fusaria autographice delineata. Annales Mycologici 15: 1–56.
- Wollenweber, H.W. & Reinking, O.A. (1935) Die Fusarien, ihre Beschreibung, Schadwirkung und Bekampfung. Verlag Paul Parey, Berlin, Germany.
- Xia, J.W., Sandoval-Denis, M., Crous, P.W., Zhang, X.G. & Lombard, L. (2019) Numbers to names–restyling the Fusarium incarnatumequiseti species complex. Persoonia 43: 186–221. https://doi.org/10.3767/persoonia.2019.43.05
- Zainudin, N.A.I.M. & Perumal, N. (2015) Mycotoxins production by Fusarium and Aspergillus species isolated from cornmeal. International Journal of Agriculture and Biology 17: 440–448. https://doi.org/10.17957/IJAB/17.3.14.170
- Zhao, L., Wei, X., Huang, C.-X., Yi, J.-P., Deng, J.-X. & Cui, M.-J. (2022) Fusarium citri-sinensis sp. nov. (Ascomycota: Nectriaceae) isolated from fruit of Citrus sinensis in China. Phytotaxa 555: 259–266. https://doi.org/10.11646/phytotaxa.555.3.5
- Zhou, X., O’Donnell, K., Aoki, T., Smith, J.A., Kasson, M.T. & Cao, Z.-M. (2016) Two novel Fusarium species that cause canker disease of prickly ash (Zanthoxylum bungeanum) in northern China form a novel clade with Fusarium torreyae. Mycologia 108: 668–681. https://doi.org/10.3852/15-189