Abstract
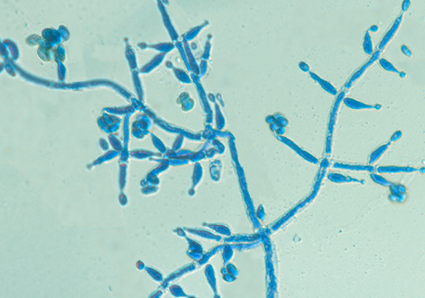
We conducted an examination of Trichoderma collections from the Wuyi Mountains in Fujian province, China. Through a comprehensive analysis that combined morphological data, culture characteristics, and phylogenetic information based on rDNA sequences of the gene encoding the second largest nuc RNA polymerase subunit (RPB2) and the partial nuc translation elongation factor 1-α encoding gene (TEF1-α), we introduce a new species called Trichoderma wuyiense, along with its asexual morphological characteristics. Trichoderma wuyiense is characterized by trichoderma- to verticillium-like conidiophores, lageniform to subulate phialides, and globose to ellipsoidal conidia. Phylogenetic analyses revealed that T. wuyiense belongs to the Viride clade and is closely related to T. sparsum. We also discuss the distinguishing features that set the new species apart from its close relatives. This study contributes to the understanding of Trichoderma diversity in the Wuyi Mountains and provides valuable insights into the morphology, phylogeny, and taxonomy of Trichoderma species. The identification of T. wuyiense adds to our knowledge of fungal biodiversity and highlights the importance of considering both morphological and molecular characteristics for accurate species classification.
References
- An, X.Y., Cheng, G.H., Gao, H.X., Li, X.F., Yang, Y., Li, D. & Li, Y. (2022) Phylogenetic analysis of Trichoderma species associated with green mold disease on mushrooms and two new pathogens on Ganoderma sichuanense. Journal of fungi 8 (7): 704. https://doi.org/10.3390/jof8070704
- Cai, F. & Druzhinina, I.S. (2021) In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Diversity 107: 1–69. https://doi.org/10.1007/s13225-020-00464-4
- Cao, Z.J., Qin, W.T., Zhao, J., Liu, Y., Wang, S.X. & Zheng, S.Y. (2022) Three new Trichoderma species in Harzianum clade associated with the contaminated substrates of edible fungi. Journal of Fungi 8 (11): 1154. https://doi.org/10.3390/jof8111154
- Chaverri, P. & Samuels, G.J. (2003) Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): Species with green ascospores. Studies in Mycology 48: 1–116.
- Chen, K. & Zhuang, W.Y. (2016) Trichoderma shennongjianum and Trichoderma tibetense, two new soil-inhabiting species in the Strictipile clade. Mycoscience 57 (5): 311–319. https://doi.org/10.1016/j.myc.2016.04.005
- Chen, K. & Zhuang, W.Y. (2017a) Seven new species of Trichoderma from soil in China. Mycosystema 36 (11): 1441–1462. https://doi.org/10.13346/j.mycosystema.170134
- Chen, K. & Zhuang, W.Y. (2017b) Discovery from a large-scaled survey of Trichoderma in soil of China. Scientific Reports 7 (1): 9090. https://doi.org/10.1038/s41598-017-07807-3
- Chen, K. & Zhuang, W.Y. (2017c) Three new soil-inhabiting species of Trichoderma in the Stromaticum clade with test of their antagonism to pathogens. Current Microbiology: 74 (9): 1049–1060. https://doi.org/10.1007/s00284-017-1282-2
- Chen, K. & Zhuang, W.Y. (2017d) Seven soil-inhabiting new species of the genus Trichoderma in the Viride clade. Phytotaxa 312 (1): 28–46. https://doi.org/10.11646/phytotaxa.312.1.2
- Chernomor, O., von Haeseler, A. & Minh, B.Q. (2016) Terrace aware data structure for phylogenomic Inference from Supermatrices. Systematic Biology 65 (6): 997–1008. https://doi.org/10.1093/sysbio/syw037
- Gams, W. & Bissett, J. (1998) Morphology and identification of Trichoderma. In Harmann, G.E. & Kubicek, C.P. (Eds.) Trichoderma and Gliocladium. Taylor and Francis, London. pp. 3–34. https://doi.org/10.1201/9781482295320
- Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. https://doi.org/10.1021/bk-1999-0734.ch008
- Hatvani, L., Vágvölgyi, C., Kredics, L. & Druzhinina, I. (2014) Chapter 3 - DNA Barcode for Species Identification in Trichoderma. In: Gupta, V.K., Schmoll, M., Herrera-Estrella, A., Upadhyay, R.S., Druzhinina, I. & Tuohy, M.G. (Eds.) Biotechnology and Biology of Trichoderma. Elsevier, Amsterdam, pp. 41–55. https://doi.org/10.1016/B978-0-444-59576-8.00003-5
- Hoang, D.T., Chernomor, O., von Haeseler, A., Minh, B.Q. & Vinh, L.S. (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. https://doi.org/10.1093/molbev/msx281
- Jaklitsch, W.M., Komon, M., Kubicek, C.P. & Druzhinina, I.S. (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97 (6): 1365–1378. https://doi.org/10.1080/15572536.2006.11832743
- Jaklitsch, W.M. & Voglmayr, H. (2013) New combinations in Trichoderma (Hypocreaceae, Hypocreales). Mycotaxon 126: 143–156. https://doi.org/10.5248/126.143
- Jaklitsch, W.M. & Voglmayr, H. (2015) Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Studies in mycology 80: 1–87. https://doi.org/10.1016/j.simyco.2014.11.001
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.F.K., von Haeseler, A. & Jermiin, L.S. (2017) Model Finder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. https://doi.org/10.1038/nmeth.4285
- Kraus, G.F., Druzhinina, I., Gams, W., Bissett, J., Zafari, D., Szakacs, G., Koptchinski, A., Prillinger, H., Zare, R. & Kubicek, C.P. (2004) Trichoderma brevicompactum sp. nov. Mycologia 96 (5): 1059–1073.
- Kredics, L., Hatvani, L., Naeimi, S., Körmöczi, P., Manczinger, L., Vágvölgyi, C. & Druzhinina, I. (2014) Chapter 1 - Biodiversity of the genus Hypocrea/Trichoderma in different habitats. In: Gupta, V.K., Schmoll, M., Herrera-Estrella, A., Upadhyay, R.S., Druzhinina, I. & Tuohy, M.G. (Ed.) Biotechnology and biology of Trichoderma. Elsevier, Amsterdam, pp. 3–24. https://doi.org/10.1016/B978-0-444-59576-8.00001-1
- Kubicek, C.P., Komon-Zelazowska, M. & Druzhinina, I.S. (2008) Fungal genus Hypocrea/Trichoderma: from barcodes to biodiversity. Journal of Zhejiang University 9 (10): 753–763. https://doi.org/10.1631/jzus.B0860015
- Li, J., Wu, Y., Chen, K., Wang, Y., Hu, J., Wei, Y. & Yang, H. (2018) Trichoderma cyanodichotomus sp. nov., a new soil-inhabiting species with a potential for biological control. Canadian journal of microbiology 64 (12): 1020–1029. https://doi.org/10.1139/cjm-2018-0224
- Liu, Y.L., Whelen, S. & Hall, B.D. (1999) Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Nguyen, L.T., Schmidt, H.A., von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. https://doi.org/10.1093/molbev/msu300
- Nirenberg, H.I. (1976) Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- Forstwirtsch 169: 1–117. https://doi.org/10.5073/20210624-085725
- Persoon, C.H. (1794) Dispositio methodica fungorum. Neues Magazin für die Botanik 1: 81–128.
- Qin, W.T., Chen, K. & Zhuang, W.Y. (2016) Five Trichoderma species new to China and notes on two other widespread species. Mycosystema 35: 994–1007. https://doi.org/10.13346/j.mycosystema.150168
- Qin, W.T. & Zhuang, W.Y. (2016a) Seven wood-inhabiting new species of the genus Trichoderma (Fungi, Ascomycota) in Viride clade. Scientific Reports 6: 27074. https://doi.org/10.1038/srep27074
- Qin, W.T. & Zhuang, W.Y. (2016b) Four new species of Trichoderma with hyaline ascospores from central China. Mycological Progress 15 (8): 811–825. https ://doi.org/10.1007/s11557-016-1211-y
- Qin, W.T. & Zhuang, W.Y. (2016c) Four new species of Trichoderma with hyaline ascospores in the Brevicompactum and Longibrachiatum clades. Mycosystema 35 (11): e160158. https://doi.org/10.13346/j.mycosystema.160158
- Qin, W.T. & Zhuang, W.Y. (2017) Seven new species of Trichoderma (Hypocreales) in the Harzianum and Strictipile clades. Phytotaxa 305 (3): 121–139. https://doi.org/10.11646/phytotaxa.305.3.1
- Qiao, M., Du, X., Zhang, Z., Xu, J. & Yu, Z. (2018) Three new species of soil-inhabiting Trichoderma from southwest China. MycoKeys (44): 63–80. https://doi.org/10.3897/mycokeys.44.30295
- Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L, Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61 (3): 539–42. https://doi.org/10.1093/sysbio/sys029
- Samuels, G.J., Dodd, S.L., Lu, B.S., Petrini, O., Schroers, H.J. & Druzhinina, I.S. (2006) The Trichoderma koningii aggregate species. Studies in mycology 56: 67–133. https://doi.org/10.3114/sim.2006.56.03
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. & Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882. https://doi.org/10.1093/nar/25.24.4876
- Zhang, C.L., Liu, S.P., Lin, F.C., Kubicek, C.P. & Druzhinina, I.S. (2007) Trichoderma taxi sp. nov., an endophytic fungus from Chinese yew Taxus mairei. FEMS microbiology letters 270 (1): 90–96. https://doi.org/10.1111/j.1574-6968.2007.00659.x
- Zhang, G.Z., Yang, H.T., Zhang, X.J., Li, J.S., Chen, K. & Huang, Y.J. (2013) Two new Chinese record of the genus Trichoderma: Trichoderma pleuroticola and T. pleurotum. Microbiology China 40 (4): 626–630.
- Zhang, G.Z., Yang, H.T., Zhang, X.J., Zhou, F.Y., Wu, X.Q., Xie, X.Y., Zhao, X.Y. & Zhou, H.Z. (2022) Five new species of Trichoderma from moist soils in China. MycoKeys 87: 133–157. https://doi.org/10.3897/mycokeys.87.76085
- Zhang, Y.B. & Zhuang, W.Y. (2019) Trichoderma bomiense and T. viridicollare, two new species forming separate terminal lineages among the green-ascospored clades of the genus. Mycosystema 38 (1): 11–22. https://doi.org/10.13346/j.mycosystema.180304
- Zhu, Z.X. & Zhuang, W.Y. (2015) Trichoderma (Hypocrea) species with green ascospores from China. Persoonia 34: 113–129. https://doi.org/10.3767/003158515X686732