Abstract
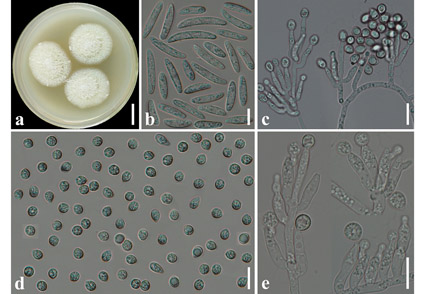
Two fungal strains were isolated from the rhizosphere soil of a Chinese medicinal herb plant, Astragalus polycladus, in northwestern Yunnan, China. Both of them were identified as a new fungal species of Varicosporellopsis based on the evidence of morphology and phylogenetic analysis of ITS, LSU, TUB and ACT sequences data. Phylogenetic analysis of combined ITS and LSU sequence data revealed that strains MTF 7 and KLF 01 belong to the genus Varicosporellopsis with the highest sequence similarity (95.51%) to the type strain V. americana CBS 148257 and formed a sister lineage to V. americana CPC 40768 with high support values (100% ML, 1.00 BI). Morphologically, V. shangrilaensis is characterized by macronematous, branched conidiophores, phialidic conidiogenous cells, reniform macroconidia and spherical microconidia. This new species is the first reported of Varicosporellopsis in China. The morphological description, illustrations and phylogenetic analysis of V. shangrilaensis are provided in this study.
References
Bellanger, J.M., Moreau, P.A., Corriol, G., Bidaud, A., Chalange, R., Dudova, Z. & Richard, F. (2015) Plunging hands into the mushroom jar: a phylogenetic framework for Lyophyllaceae (Agaricales, Basidiomycota). Genetica 143: 169–194. https://doi.org/10.1007/s10709-015-9823-8
Bills, G.F., Platas, G., Overy, D.P., Collado, J., Fillola, A., Jiménez, M.R., Martín, J., del Val, A.G., Vicente, F., Tromo, J.R., Peláez, F., Calati, K., Harris, G., Parish, C., Xu, D.M. & Roemer, T. (2009) Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia 101: 449–472. https://doi.org/10.3852/08-163
Brock, T.D., Madigan, M.T., Martinko, J.M. & Parker, J. (2003) Brock biology of microorganisms. Upper Saddle River, Prentice Hall.
Bunyard, B.A., Nicholson, M.S., Royse, D. (1994) A systematic assessment of Morchella using RFLP analysis of the 28S ribosomal RNA gene. Mycologia 86: 762–772. https://doi.org/10.2307/3760589
Cabral, A., Rego, C., Nascimento, T., Oliveira, H., Groenewald, J.Z. & Crous, P.W. (2012) Multi-gene analysis and morphology reveal novel Ilyonectria species associated with black foot disease of grapevines. Fungal Biology 116: 62–80. https://doi.org/10.1016/j.funbio.2011.09.010
Chaverri, P., Salgado, C., Hirooka, Y., Rossman, A. & Samuels, G. (2011) Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Studies in Mycology 68: 57–78. https://doi.org/10.3114/sim.2011.68.03
Chen, C., Gong, G.F., Liang, J.Y., Dou, Z.R., Ding, J.J., Jiang, B. & Wang, K.L. (2022) Rhizosphere microbial diversity of Astragalus yunnanensis and A. tatsienensis var. incanus in Baima Snow Mountain and screening of microorganisms with anti-biofilm activity. Microbiology China 49: 3813–3836 https://doi.org/10.13344/j.microbiol.china.220055
Crous, P.W., Osieck, E.R., Jurjevi, Ž., Boers, J., Van Iperen, A.L., Starink-Willemse, M., Dima, B., Balashov, S., Bulgakov, T.S., Johnston, P.R., Morozova, O.V., Pinruan, U., Sommai, S., Alvarado, P., Decock, C.A., Lebel, T., McMullan-Fisher, S., Moreno, G., Shivas, R.G., Zhao, L., Abdollahzadeh, J., Abrinbana, M., Ageev, D.V., Akhmetova, G., Alexandrova, A.V., Altés, A., Amaral, A.G.G., Angelini, C., Antonín, V., Arenas, F., Asselman, P., Badali, F., Baghela, A., Bañares, A., Barreto, R.W., Baseia, I.G., Bellanger, J.M., Berraf-Tebbal, A., Biketova, A.Y., Bukharova, N.V., Burgess, T.I., Cabero, J., Câmara, M.P.S., Cano-Lira, J.F., Ceryngier, P., Chávez, R., Cowan, D.A., de Lima, A.F., Oliveira, R.L., Denman, S., Dang, Q.N., Dovana, F., Duarte, I.G., Eichmeier, A., Erhard, A., Esteve-Raventós, F., Fellin, A., Ferisin, G., Ferreira, R.J., Ferrer, A., Finy, P., Gaya, E., Geering, A.D.W., Gil-Durán, C., Glässnerová, K., Glushakova, A.M., Gramaje, D., Guard, F.E., Guarnizo, A.L., Haelewaters, D., Halling, R.E., Hill, R., Hirooka, Y., Hubka, V., Iliushin, V.A., Ivanova, D.D., Ivanushkina, N.E., Jangsantear, P., Justo, A., Kachalkin, A.V., Kato, S., Khamsuntorn, P., Kirtsideli, I.Y., Knapp, D.G., Kochkina, G.A., Koukol, O., Kovács, G.M., Kruse, J., Kumar, T.K.A., Kušan, I., Læssøe, T., Larsson, E., Lebeuf, R., Levicán, G., Loizides, M., Marinho, P., Luangsa-ard, J.J., Lukina, E.G., Magaña-Dueñas, V., Maggs-Kölling, G., Malysheva, E.F., Malysheva, V.F., Martín, B., Martín, M.P., Matočec, N., McTaggart, A.R., Mehrabi-Koushki, M., Mešić, A., Miller, A.N., Mironova, P., Moreau, P.A., Morte, A., Müller, K., Nagy, L.G., Nanu, S., Navarro-Ródenas, A., Nel, W.J., Nguyen, T.H., Nóbrega, T.F., Noordeloos, M.E., Olariaga, I., Overton, B.E., Ozerskaya, S.M., Palani, P., Pancorbo, F., Papp, V., Pawłowska, J., Pham, T.Q., Phosri, C., Popov, E.S., Portugal, A., Pošta, A., Reschke, K., Reul, M., Ricci, G.M., Rodríguez, A., Romanowski, J., Ruchikachorn, N., Saar, I., Safi, A., Sakolrak, B., Salzmann, F., Sandoval-Denis, M., Sangwichein, E., Sanhueza, L., Sato, T., Sastoque, A., Senn-Irlet, B., Shibata, A., Siepe, K., Somrithipol, S., Spetik, M., Sridhar, P., Stchigel, A.M., Stuskova, K., Suwannasai, N., Tan, Y.P., Thangavel, R., Tiago, I., Tiwari, S., Tkalčec, Z., Tomashevskaya, M.A., Tonegawa, C., Tran, H.X., Tran, N.T., Trovão, J., Trubitsyn, V.E., Van Wyk, J., Vieira, W.A.S., Vila, J., Visagie, C.M., Vizzini, A., Volobuev, S.V., Vu, D.T., Wangsawat, N., Yaguchi, T., Ercole, E., Ferreira, B.W., de Souza, A.P., Vieira, B.S. & Groenewald, J.Z. (2021) Fungal Planet description sheets: 1284–1382. Persoonia - Molecular Phylogeny and Evolution of Fungi 47: 178–374. https://doi.org/10.3767/persoonia.2021.47.06
Crous, P.W., Schumacher, R.K., Wingfield, M.J., Akulov, A., Denman, S., Roux, J., Braun, U., Burgess, T.I., Carnegie, A.J., Vaczy, K.Z., Guatimosim, E., Schwartsburd, P.B., Barreto, R.W., Hernandez-Restrepo, M., Lombard, L. & Groenewald, J.Z. (2018) New and Interesting Fungi. 1. Fungal Systematics and Evolution 1: 169–216. https://doi.org/10.3114/fuse.2018.01.08
Felsenstein, J. (1985) Confidence-limits on phylogenies: anapproach using the bootstrap. Evolution 39: 783–791. https://doi.org/10.2307/2408678
Groenewald, J.Z., Nakashima, C., Nishikawa, J., Shin, H.D., Park, J.H., Jama, A.N., Groenewald, M., Braun, U. & Crous, P.W. (2013) Species concepts in Cercospora: spotting the weeds among the roses. Studies in Mycology 75 (1): 115–170. https://doi.org/10.3114/sim0012
Hall, B.G. (2007) Phylogenetic trees made easy: a how-to manual. Massachusetts: Sinauer Associates Sunderland: Sunderland, MA, USA.
Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
Herrera, C.S., Rossman, A., Samuels, G., Lechat, C. & Chaverri, P. (2013a) Revision of the genus Corallomycetella with Corallonectria gen. nov. for C. jatrophae (Nectriaceae, Hypocreales). Mycosystema 32: 518–544
Herrera, C.S., Rossman, A.Y., Samuels, G.J. & Chaverri, P. (2013b) Pseudocosmospora, a new genus to accommodate Cosmospora vilior and related species. Mycologia 105: 1287–1305. https://doi.org/10.3852/12-395
Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Joshi, Y., Bansal, P. & Yadav, A.L. (2022) Cercidospora navarroi, a new species of lichenicolous fungus from the Central Himalayan region of India. Phytotaxa 549: 241–246. https://doi.org/10.11646/phytotaxa.549.2.10
Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010
Lechat, C. & Fournier, J. (2016) Varicosporellopsis, a new aquatic genus from southern France. Ascomyceteorg 8: 96–100.
Li, Y., Yang, Y., Zhang, H., Wei, G. & Li, Z. (2021) Rhizosphere bacterial and fungal spatial distribution and network pattern of Astragalus mongholicus in representative planting sites differ the bulk soil. Applied Soil Ecology 168: 104114. https://doi.org/10.1016/j.apsoil.2021.104114
Liu, Q.L., Li, J.Q., Wingfield, M.J., Duong, T.A., Wingfield, B.D., Crous, P.W. & Chen, S.F. (2020) Reconsideration of species boundaries and proposed DNA barcodes for Calonectria. Studies in Mycology 97 (1): 100106–100106. https://doi.org/10.1016/j.simyco.2020.08.001
Lombard, L., van der Merwe, N.A., Groenewald, J.Z. & Crous, P.W. (2015) Generic concepts in Nectriaceae. Studies in Mycology 80: 189–245. https://doi.org/10.1016/j.simyco.2014.12.002
Marasas, W.F.O., Nelson, P.E. & Toussoun, T.A. (1984) Toxigenic Fusarium species: identity and mycotoxicology. Pennsylvania State University press, University, PA.
Ming, D.Q., Luo, L.Y., He, X.X., Wang, M.S., Fang, W.X., Chen, S.F., Chen, W.H., Han, Y.F. & Liang, Z.Q. (2021) Paracremonium lepidopterorum, a new insect-associated fungus. Phytotaxa 524: 85–91. https://doi.org/10.11646/phytotaxa.524.2.2
Nguyen, L.T., Schmidt, H.A., Von Haeseler, A. & Minh, B.Q. (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. https://doi.org/10.1093/molbev/msu300
Nguyen, T.T., Voigt, K., Santiago, A.L.C.M.d.A., Kirk, P.M. & Lee, H.B. (2021) Discovery of novel Backusella (Backusellaceae, Mucorales) isolated from invertebrates and toads in Cheongyang, Korea. Journal of Fungi 7: 513. https://doi.org/10.3390/jof7070513
Posada, D. (2008) jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. https://doi.org/10.1093/molbev/msn083
Rambaut, A. (2010) FigTree v1. 3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh.
Rossman, A.Y. (1996) Morphological and molecular perspectives on systematics of the Hypocreales. Mycologia 88: 1–19. https://doi.org/10.2307/3760780
Rossman, A.Y., Samuels, G.J., Rogerson, C.T. & Lowen, R. (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1–248.
Summerbell, R. (2003) Aspergillus, Fusarium, Sporothrix, Piedraia, and their relatives. In: Howard, D.H. (ed.) Pathogenic Fungi in Humans and Animals. Marcel Dekker Press, New York. pp. 237–498.
Schroers, H.J., Geldenhuis, M., Wingfield, M., Schoeman, M., Yen, Y.F., Shen, W.C. & Wingfield, B. (2005) Classification of the guava wilt fungus Myxosporium psidii, the palm pathogen Gliocladium vermoesenii and the persimmon wilt fungus Acremonium diospyri in Nalanthamala. Mycologia 97: 375–395. https://doi.org/0.3852/mycologia.97.2.375
Schroers, H.J., Gräfenhan, T., Nirenberg, H. & Seifert, K. (2006) A revision of Cyanonectria and Geejayessia gen. nov., and related species with Fusarium-like anamorphs. Studies in Mycology 56: 178–179. https://doi.org/10.3114/sim.2011.68.05
Sun, B.D., Zhou, Y.G. & Chen, A.J. (2017) Bisifusarium tonghuanum (Nectriaceae), a novel species of Fusarium-like fungi from two desert oasis plants. Phytotaxa 317: 123–129. https://doi.org/10.11646/phytotaxa.317.2.4
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. https://doi.org/10.1093/molbev/mst197
Vu, D., Groenewald, M., de Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., Al-Hatmi, A., Groenewald, J.Z., Cardinali, G., Houbraken, J., Boekhout, T., Crous, P.W., Robert, V. & Verkley, G.J.M. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. https://doi.org/10.1016/j.simyco.2018.05.001
White, T.J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 18: 315–322.
Wijayawardene, N.N., Phillips, A.J.L., Pereira, D.S., Dai, D.Q., Aptroot, A., Monteiro, J.S., Druzhinina, I.S., Cai, F., Fan, X.L., Selbmann, L., Coleine, C., Castaneda-Ruiz, R.F., Kukwa, M., Flakus, A., Fiuza, P.O., Kirk, P.M., Kumar, K.C.R., Arachchi, I.S.I., Suwannarach, N., Tang, L.Z., Boekhout, T., Tan, C.S., Jayasinghe, R. & Thines, M. (2022) Forecasting the number of species of asexually reproducing fungi (Ascomycota and Basidiomycota). Fungal Diversity 114: 463–490. https://doi.org/10.1007/s13225-022-00500-5
Zeng, Z.Q. & Zhuang, W.Y. (2022) New Species of Nectriaceae (Hypocreales) from China. Journal of Fungi 8. https://doi.org/10.3390/jof8101075
Zhang, F., Liu, S.R., Zhou, X.J., Monkai, J., Hongsanan, S., Shang, Q.J., Hyde, K.D. & Yang, X.Y. (2020) Fusarium xiangyunensis (Nectriaceae), a remarkable new species of nematophagous fungi from Yunnan, China. Phytotaxa 450: 273–284. https://doi.org/10.11646/phytotaxa.450.3.3