Abstract
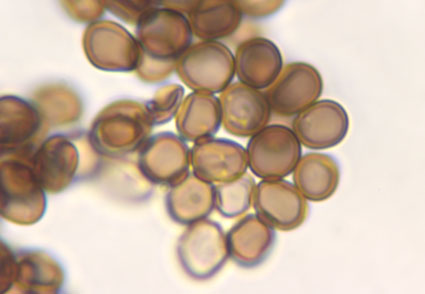
The morphology and phylogeny of Tranzscheliella on Agropyron cristatum (Triticeae) collected in Ukraine were studied by light microscopy, scanning electron microscopy and a molecular phylogenetic data of two rDNA loci (ITS and 28S). As a result, T. helutae sp. nov. is described. The fungus differs from other species from the T. hypodytes complex by a combination of morphological and molecular data.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Begerow, D., Stoll, M. & Bauer, R. (2006) A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia 98: 906–916. https://doi.org/10.1080/15572536.2006.11832620</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Castresana, J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Gams, W., Stalpers, J.A., Robert, V. & Stegehuis, G. (2004) MycoBank: an online initiative to launch mycology into the 21<sup>st</sup> century. Studies in Mycology 50: 19–22.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dewey, D.R. (1986) Taxonomy of the crested wheatgrass (Agropyron). In: Johnson, K.L. (ed.) Crested wheatgrass: its values, problems and myths. Utah State University, Logan, pp. 31–44. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Fischer, G.W. & Hirschhorn, E. (1945) A critical study of some species of Ustilago causing stem smut of various grasses. Mycologia 37: 236–266. https://doi.org/10.1080/00275514.1945.12023984</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Gatesy, J., Desalle, R. & Wheeler, W.C. (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Molecular Phylogenetics and Evolution 2: 152–157. https://doi.org/10.1006/mpev.1993.1015</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Giribet, G. & Wheeler, W.C. (1999) On gaps. Molecular Phylogenetics and Evolution 13: 132–143. https://doi.org/10.1006/mpev.1999.0643</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Lavrov, N. (1936) Ustilaginacese novae el rare Asiae borealis centralisque. Trudy Biologicheskiye Naucno-Issledovatelskogo Instituta Tomskogo Gosudarstvennogo Universiteta 2: 1–35. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Li, Y-M., Shivas, R.G. & Cai, L. (2017) Cryptic diversity of Tranzscheliella spp. (Ustilaginales) is driven by host switches. Nature Scientific Reports 7: 43549. https://doi.org/10.1038/srep43549</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">McTaggart, A.R., Prychild, C.J., Bruhl, J.J. & Shivas, R.G. (2020) The PhyloCode applied to Cintractiellales, a new order of smut fungi with unresolved phylogenetic relationships in the Ustilaginomycotina. Fungal Systematics and Evolution 6 (1): 55–64. https://doi.org/10.3114/fuse.2020.06.04</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nylander, J.A., Wilgenbusch, J.C., Warren, D.L. & Swofford, D.L. (2008) AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. https://doi.org/10.1093/bioinformatics/btm388</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">O’Donnell, K. (1993) Fusarium and its near relatives. In: Reynolds, D.R. & Taylor, J.W. (eds.) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp. 225–233.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Pi?tek, M., Lutz, M., Sousa, F.M.P., Santos, A.R.O., Felix, C.R., Landell, M.F., Gomes, F.C.O. & Rosa, C.A. (2017) Pattersoniomyces tillandsiae gen. et comb. nov.: linking sexual and asexual morphs of the only known smut fungus associated with Bromeliaceae. Organismal Diversity and Evolution 17: 531–543. https://doi.org/10.1007/s13127-017-0340-8</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A. (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stoll, M., Piepenbring, M., Begerow, D. & Oberwinkler, F. (2003) Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Canadian Journal of Botany 81: 976–984. https://doi.org/10.1139/b03-094</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38: 3022–3027. https://doi.org/10.1093/molbev/msab120</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2003) Taxonomical studies on Ustilaginales. XXIII. Mycotaxon 85: 1–65. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2003b) The smut fungi (Ustilaginomycetes) of Sporobolus (Poaceae). Fungal Diversity 14: 205–241. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2012) Smut fungi of the world. APS Press, St. Paul, MN, 1458 pp.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2013) Illustrated genera of smut fungi 3<sup>rd</sup> edition. APS Press, St. Paul, MN. https://doi.org/10.5943/mycosphere/4/3/2</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. & Mckenzie, E.H. (2002) Smut fungi of New Zealand. Fungal Diversity Press, University of Hong Kong.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Castresana, J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Gams, W., Stalpers, J.A., Robert, V. & Stegehuis, G. (2004) MycoBank: an online initiative to launch mycology into the 21<sup>st</sup> century. Studies in Mycology 50: 19–22.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dewey, D.R. (1986) Taxonomy of the crested wheatgrass (Agropyron). In: Johnson, K.L. (ed.) Crested wheatgrass: its values, problems and myths. Utah State University, Logan, pp. 31–44. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Fischer, G.W. & Hirschhorn, E. (1945) A critical study of some species of Ustilago causing stem smut of various grasses. Mycologia 37: 236–266. https://doi.org/10.1080/00275514.1945.12023984</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Gatesy, J., Desalle, R. & Wheeler, W.C. (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Molecular Phylogenetics and Evolution 2: 152–157. https://doi.org/10.1006/mpev.1993.1015</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Giribet, G. & Wheeler, W.C. (1999) On gaps. Molecular Phylogenetics and Evolution 13: 132–143. https://doi.org/10.1006/mpev.1999.0643</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Lavrov, N. (1936) Ustilaginacese novae el rare Asiae borealis centralisque. Trudy Biologicheskiye Naucno-Issledovatelskogo Instituta Tomskogo Gosudarstvennogo Universiteta 2: 1–35. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Li, Y-M., Shivas, R.G. & Cai, L. (2017) Cryptic diversity of Tranzscheliella spp. (Ustilaginales) is driven by host switches. Nature Scientific Reports 7: 43549. https://doi.org/10.1038/srep43549</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">McTaggart, A.R., Prychild, C.J., Bruhl, J.J. & Shivas, R.G. (2020) The PhyloCode applied to Cintractiellales, a new order of smut fungi with unresolved phylogenetic relationships in the Ustilaginomycotina. Fungal Systematics and Evolution 6 (1): 55–64. https://doi.org/10.3114/fuse.2020.06.04</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nylander, J.A., Wilgenbusch, J.C., Warren, D.L. & Swofford, D.L. (2008) AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24: 581–583. https://doi.org/10.1093/bioinformatics/btm388</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">O’Donnell, K. (1993) Fusarium and its near relatives. In: Reynolds, D.R. & Taylor, J.W. (eds.) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp. 225–233.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Pi?tek, M., Lutz, M., Sousa, F.M.P., Santos, A.R.O., Felix, C.R., Landell, M.F., Gomes, F.C.O. & Rosa, C.A. (2017) Pattersoniomyces tillandsiae gen. et comb. nov.: linking sexual and asexual morphs of the only known smut fungus associated with Bromeliaceae. Organismal Diversity and Evolution 17: 531–543. https://doi.org/10.1007/s13127-017-0340-8</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A. (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stoll, M., Piepenbring, M., Begerow, D. & Oberwinkler, F. (2003) Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Canadian Journal of Botany 81: 976–984. https://doi.org/10.1139/b03-094</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tamura, K., Stecher, G. & Kumar, S. (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution 38: 3022–3027. https://doi.org/10.1093/molbev/msab120</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2003) Taxonomical studies on Ustilaginales. XXIII. Mycotaxon 85: 1–65. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2003b) The smut fungi (Ustilaginomycetes) of Sporobolus (Poaceae). Fungal Diversity 14: 205–241. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2012) Smut fungi of the world. APS Press, St. Paul, MN, 1458 pp.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. (2013) Illustrated genera of smut fungi 3<sup>rd</sup> edition. APS Press, St. Paul, MN. https://doi.org/10.5943/mycosphere/4/3/2</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vánky, K. & Mckenzie, E.H. (2002) Smut fungi of New Zealand. Fungal Diversity Press, University of Hong Kong.</span></span></span>